In The News

06 October 2025
Stowers Investigator Named "20 to Know" in Kansas City
From the Kansas City Business Journal, Investigator Julia Zeitlinger named "20 to Know"
Read Article
By Alissa Poh
Growing up in Germany, Associate Investigator Julia Zeitlinger, PhD, excelled in math and science in school with many seeing traits that would fit naturally with a scientific career: curiosity, a liking for theory construction and an analytical mind. But her decision to become a scientist was not without hesitation. She also loved the arts and has fond memories of stepping out onto a stage.
“Everything finally comes together on opening night; there’s a deep quiet in the audience as lines are spoken and listened to, and you’re really living in the moment,” says Zeitlinger. “Sometimes I miss that electric atmosphere.”
Ultimately, she fell in love with biology in college and her first stint at laboratory research at King’s College London kindled a long-lasting spark that went far beyond the allure of opening night. On the heels of a PhD in developmental genetics from the European Molecular Biology Laboratory in Heidelberg, Zeitlinger pursued a postdoctoral fellowship at the Massachusetts Institute of Technology.
“The first microarray publications were just out and genomics was coming into its own; I wanted to be where cutting-edge discoveries were being made,” she recalls. “I was intrigued by the prospects of probing the black box of transcription programs, and spending a few years in a different country.”
Zeitlinger never intended to stay past those few years, but she fell in love again—only this time she got married and decided to make the US her permanent home. “Life takes unexpected twists and turns and it’s important to recognize and seize the wonderful opportunities that present themselves when you least expect it,” and adds with a laugh, “not unlike research.”
Since Zeitlinger joined the Stowers Institute in 2007, she is ferreting out, in Drosophila, fresh threads linking DNA sequence with cellular function. In particular, she and her team have taken a closer look at promoters and learned that there’s more to these DNA regions, responsible for initiating transcription of genes into messenger RNAs, than meets the scientific eye. They dictate, for instance, whether RNA polymerases, or the primary workhorses of transcription, face nonstop action or have the flexibility of pausing on the job. Varying developmental needs drive these and other behavioral cues, and promoters have all the necessary details.
“Promoters have often been dismissed as boring from a research standpoint,” Zeitlinger says. “But we’re finding otherwise. Basically, transcription isn’t just this behemoth of proteins doing one job the same way.”
What advice would you offer someone contemplating a career in science while also toying with the arts, given that ne’er the twain shall meet, professionally speaking?
I’d start by emphasizing that one is not somehow less creative than the other. Yes, creativity in science is often more hidden, especially at the beginning when you’re establishing your own research niche and are swamped with experiments that seem tedious and endless. But it’s a lot like artists with their first sketches—they’re trying out a concept to see if it works visually, and if it doesn’t, they come up with modifications. The final painting is not immediately evident. In science, the beauty is when things go in unexpected directions; if you’re open to change, you’ll find that’s where not only surprises but the really important discoveries often lurk.
In the end, I would say follow your heart and do what you’re most passionate about. Science can be a tough business of coping with failed experiments, confusing results and criticism from peers. Having a place within yourself to which you can always return, when the going gets rough, is key. I never felt that I had that confidence with acting, as much as I loved the stage.
Regarding your research, what’s the most exciting question you’re planning to tackle this year?
I hope we come closer to solving what’s known as the combinatorial code for transcription: basically, all the factors and signals that come together to turn on a gene. There’s much we don’t yet understand about this switchboard’s manual. Which are the toggles to press, and in what order? To gain new insights, we’ve recently developed a technology called ChIP-nexus that allows us to zoom in at much higher resolution on not only DNA regions where transcription factors are found but on their precise binding sites in the genome. We can already see interesting results but the big challenge will be to find out how these pieces fit into the big puzzle.
We’re also exploring the timing, or speed, of embryonic development in flies. It’s thought of as a program with set steps, but it occurs at surprisingly different rates across the animal kingdom. Even among flies it is remarkably different – a mere 14 hours in Drosophila yakuba, for instance; and 32 hours in Drosophila mojavensis. Our measurements also show that the mean embryonic development times for different lines from a single Drosophila melanogaster population range from 18 to 23 hours. So genetic variation among individuals has its own impact here, yet we have no idea what mechanisms are at play. We’ve established a nice system that will allow us to hunt for and examine factors influencing these differences.
What are your thoughts on the encode (encyclopedia of dna elements) project’s overall applicability and importance?
I believe it’s an important step toward understanding how genome information encodes cellular function and I hope it will make genome-centric research more mainstream. Genome-wide experiments are still viewed as a starting point for more detailed single-gene analyses. But I believe genomics should be a discipline in its own right, for it has its own methods of extracting information and testing hypotheses. The power lies in statistics on a genome-wide scale – with careful interpretation of correlations and P values, of course. For instance, if I’d first observed RNA polymerase II’s pausing behavior through a single-gene analysis rather than genome-wide, I might have regarded the gene itself as weird or incorrectly mapped, rather than believe what I saw was real.
That said, at least some of what the public first heard about ENCODE in 2013 was oversimplified. This project contains huge, complex data sets, yet scientists had to come up with a simple punch line, which resulted in statements like “Junk DNA is not really junk.” I prefer Eric Lander’s seven-word summary in 2003: “Genome: bought the book; hard to read.” Despite all the progress, it’s still true.
How do you feel about scientists cherry-picking data to report – or omit, as the case may be – which occurs even more often than outright fraud?
I’m not sure trying to present the best or “nicest” data is necessarily cherry-picking, as long as you can show that your findings are real, reproducible and accompanied by robust statistics. Distinguishing between actual results and interpretation is key, however: the first needs to be solid, while the second can change.
There’s also this tendency to start with just one hypothesis, for which confirmatory evidence is sought. As Karl Popper pointed out fifty years ago, that’s not good science. It’s critical to have and test multiple hypotheses; rejecting one doesn’t mean any of the rest are correct, either. There could be other possibilities you might not have envisioned at the time.
Where do you do your best thinking?
For me, it’s when more than where: in the morning, right after waking up. It’s quiet and my eyes are still mostly closed; somehow, thoughts and ideas that were swirling around in my mind have crystallized overnight.
In general, is saying no difficult or easy for you?
I used to be quite bad at it, until I read William Ury’s The Power of a Positive No. It really woke me up to the importance of accounting for my own core values and priorities. If something isn’t worthwhile to me, I can now say no and, as a pretty direct person at heart, I feel much better being frank. But it didn’t come naturally.
What historical period would you most like to visit, if it were possible?
I can think of two. For my son’s sake, it would be great to scope out the Jurassic period and see actual dinosaur colors, as well as some of the earliest birds like Archaeopteryx. My own great interest is human evolution, so I’d choose a visit with the Neanderthals to get a better idea of just how similar to modern humans they were, how they communicated, and why they were eventually outcompeted.
What does your perfect day look like?
I had one last weekend, actually – half a day at work, where I was able to do some reading to catch up on
topics of interest, before spending the rest of it with my family. Having a three- and five-year-old is definitely
hectic, but we’re making beautiful memories together and I wouldn’t
trade my life right now for anything else.
In The News

06 October 2025
From the Kansas City Business Journal, Investigator Julia Zeitlinger named "20 to Know"
Read Article
News
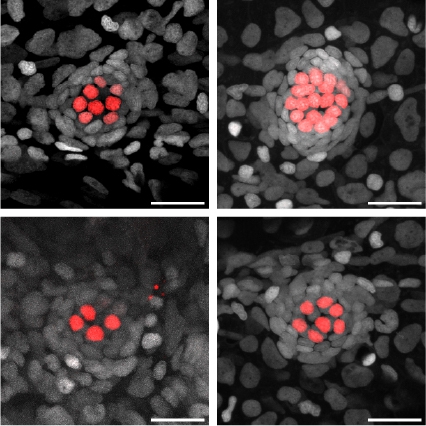
03 October 2025
Stowers scientists discover a master gene in zebrafish that determines hair cell type, a potential target for restoring hearing loss in humans
Read Article
In The News
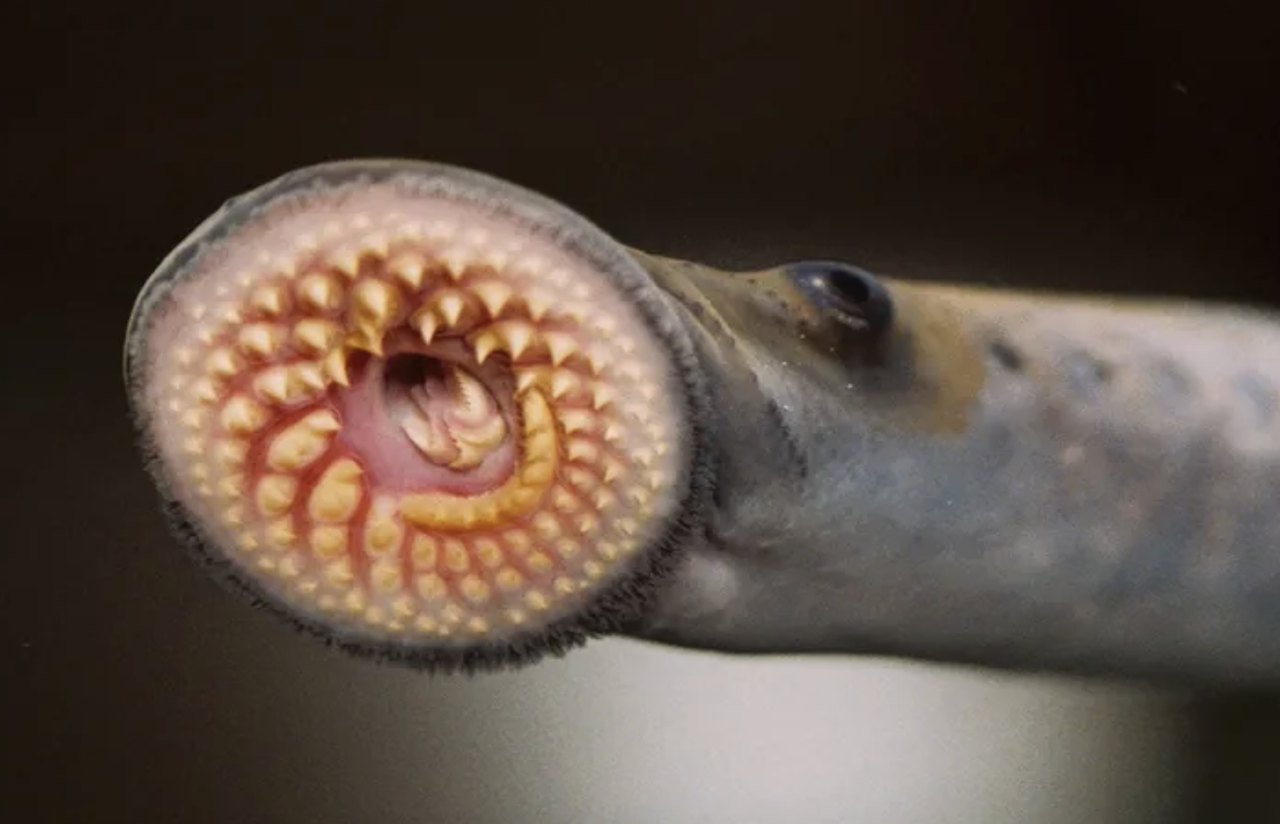
30 September 2025
From Smithsonian Magazine, The sea lamprey looks like it’s from another planet, but this ancient creature has a surprising amount in common with humans.
Read Article
