

Apr 05-092025
Kansas City, MO
The 18th Annual International Biocuration Conference provides a forum for curators, developers, and users of clinical and life sciences data, knowledge, and models to discuss their work, promote collaboration, and foster the community around this active and growing area of research. Participants from academia, government, and industry interested in the tools, methodology, and philosophy of curation in the clinical and life sciences are encouraged to attend and submit an abstract for consideration for an oral or poster presentation. This conference is organized in part with the International Society of Biocuration (ISB).
Share your experience at the conference, connect with other attendees, and learn more by using the hashtag #Biocuration2025!
Keynote Speakers
Registration is now open!
Please contact biocuration2025@stowers.org with any queries relating to the registration process.
Register here
Key dates:
Workshops: April 5-6, 2025
Conference: April 7-9, 2025
April 9: Biocuration careers workshop
Registration:
Registration opens: September 3, 2024
Early Bird Registration: September 3, 2024 - December 2, 2024
Registration closes: March 28, 2025 (or when attendee cap is met)
Abstracts:
Submission: September 3, 2024 - November 14, 2024
Talk and poster decision notification: December 1-5, 2024
Registration Information
The 18th International Biocuration Conference is organized in collaboration with the International Society for Biocuration (ISB). Members save significantly on the registration fee. Click here for more information about ISB membership.
The registration fee covers:
- Attendance at all sessions, including workshops
- Continental breakfast on full conference days
- Lunch on full conference days
- Coffee/refreshments during breaks
- Poster receptions
Please note that travel and lodging is not included.
Registration Types and Pricing:
Academic ISB member: $400 ($450 after 12/2)
Academic Non-member: $525 ($575 after 12/2)
Non-academic ISB member: $575 ($625 after 12/2)
Non-academic Non-member: $700 ($775 after 12/2)
Student/retiree/low-income countries ISB member: $200 ($250 after 12/2)
Student/retiree/low-income countries Non-member: $325 ($375 after 12/2)
Virtual ISB member: $150 ($250 after 12/2)
Virtual Non-member: $250 ($350 after 12/2)
Registration closes: March 28, 2025 (or when attendee cap is met)
Conference Dinner
An off-site social dinner will be held on the night of Tuesday, April 8, 2025 at 7:00 PM. This will include Kansas City-style barbecue as well as purposeful vegetarian, vegan, and gluten-free options. $85.
Travel Grants
Travel grants are available from the International Society of Biocuration (ISB). More information on eligibility and application can be found here. The application period closes on November 25, 2024.
Abstract submission
Everyone is welcome to submit an abstract. During registration, participants will have the opportunity to submit an abstract and indicate if they want to be considered for a talk or poster.
Abstract Deadline: November 14, 2024
Participants with abstracts that are selected for posters and talks will be notified during the first week of December.
Biocuration Careers Workshop
At the conference we will hold a session promoting careers in the Biocuration field. We welcome any students, trainees, post-docs or staff from the local Kansas City area to participate in the workshop and learn more about job opportunities in this field and learn more about the field. You do not have to register for the conference to attend. Learn more at the registration link below.
Register hereWorkshops
Biocuration 2025 will be hosting 13 workshops at the main conference venue. While most precede the main conference, the Biocuration Career Opportunities workshop hosted by the ISB will be held during the main agenda of the final day of the conference. Here’s a quick look:
Empowering biocurators with APICURON: from curators’ needs to individual profiles development and integration of ontologies and datasets
Saturday: 9am - 12:30pm, Organized by Silvio Tosatto
This half-day workshop explores APICURON’s comprehensive solutions for biocurators, covering curators’ needs, developing individual profiles, and integrating US-based biomedical ontologies, curated resources, and datasets. Tailored for curators worldwide, this event is especially beneficial for those biocurators seeking to enhance their opportunities, including curators from countries with limited funding.
SESSION DURATION DESCRIPTION
Welcome and Introduction: 15 minutes. The workshop will begin with a brief introduction to APICURON, providing an overview of its current scope and outlining the workshop’s objectives.
Overview of APICURON: 30 minutes. This session will explore the details of APICURON’s functionalities. Participants will learn about the success stories and use cases of current APICURON partners, providing a solid foundation for understanding how APICURON operates and its benefits.
Addressing the needs of biocurators: 45 minutes. Identifying and addressing the specific needs of curators, including those from low- and lower-middle income countries and with limited funding. Highlight the challenges faced by curators in different regions and discuss how APICURON can evolve to meet these diverse needs. Explore strategies for APICURON to support a diverse range of curators.
Individual profiles for biocurators: 40 minutes. The development of individual APICURON profiles will be introduced, showcasing the benefits for freelance curators and those working on assigned projects (e.g. curators employed in curation companies). Individual biocurator profiles will be particularly beneficial also for curators working in low and lower-middle income countries, where funding may be limited but many skilled curators are present.
Integrating manually curated datasets and biomedical ontologies: 40 minutes. This session will discuss the integration of manually curated datasets in APICURON as well as biomedical ontologies. Participants will explore examples of biomedical ontologies and curated datasets that could be integrated and consider the technical aspects of this integration.
Integration of resources: expanding to the US and beyond: 40 minutes. This session will identify key US-based manually curated biological databases, biomedical ontologies, manually curated datasets that are interested in partnering with APICURON. Representatives from US-based databases will be invited to establish new collaborations with APICURON.
Future developments and closing remarks: 25 minutes. The final session will summarize the key points discussed throughout the workshop as well as outline next steps and follow-up actions.
Collaborative Biocuration Workflows: Lessons learned from the OBO Academy
Saturday: 9am - 12:30pm, Organized by Sabrina Toro and Nicole Vasilevsky
Collaborative open science is crucial for advancing and maximizing the impact of biocuration. It has been practiced in the development of ontologies, as described in the OBO Academy training materials. In this workshop, we will explore best practices for open science, focusing on contributions to open ontology development and biocuration. We will discuss the lessons learned from open ontology development, leveraging the freely available resources provided by the OBO Academy, and explore how these can be applied to biocuration more generally.
Our workshop will address open science best practices in two key areas: 1) open biomedical and biological ontology development, and 2) open biocuration projects. We will cover best practices for contributing to Open Biomedical and Biological Ontology (OBO) Foundry ontologies. Many biocurators rely on OBO Foundry ontologies for data standardization and annotation. These ontologies are developed and maintained by volunteers or distributed teams and are open for community contributions. Through real-life case studies and an introduction to current tools, we will explore how to utilize GitHub for issue tracking, documentation, discussions, and automation pipelines, and examine social workflows and adherence to open science principles.
In addition, we will discuss strategies for maintaining and advancing community-based projects, emphasizing effective collaboration and contributions to open-source initiatives. We will invite speakers from successful community-sourced curation projects.
Websites:
OBO Foundry: https://obofoundry.org/
OBO Academy: https://oboacademy.github.io/obook/
Workshop duration: Half-day
Apollo 3 and JBrowse 2 Setup and Usage
Saturday 10:30 am - 12:30 pm, Organized by Garrett Stevens and Colin Diesh
Apollo 3 is a genomic annotation editor, built on the JBrowse 2 genome browser, that supports collaborative editing with real-time updates of shared annotations. Apollo 3 is designed to be useful for both small scale projects (e.g. easy usage in JBrowse Desktop without complex database setup) and large scale projects (e.g. fine grained authorization and authentication, user defined scripts for data flagging and validation, and more). Apollo 3 takes advantage of features like the JBrowse 2 synteny view, allowing users to interactively edit and compare annotations across species. In this workshop, we will provide an interactive tutorial to demonstrate setup and usage of Apollo 3 and JBrowse 2, and demonstrate newly developed features for visualizing protein structures and multiple sequence alignments with connections to AlphaFoldDB.
Enhancing the Unified Phenotype Ontology to Support Cross-Species Phenotype Interoperability
Saturday: 1:30 pm - 5 pm, Organized by Susan Bello and Arwa Ibrahim
Workshop goal: Advance community efforts to curate and compare phenotypes within and across species, bringing together the Unified Phenotype Ontology (uPheno) user community to improve usability and applicability.
The Unified Phenotype Ontology (uPheno) project began with the goal to bring together species neutral phenotype ontology to support cross-species presentation and analysis of phenotypes. Much work has been done to develop methods to bring together disparate organism specific phenotype ontologies. A suite of patterns has been developed to create consistent logical definitions for terms across phenotype ontologies, decreasing the variation between these organism-specific ontologies, allowing for generation of new terms, and facilitating automated integration into uPheno. Manual and automated mapping efforts of terms using the Simple Standard for Sharing Ontology Mappings (SSSOM) framework augment the pattern-based integration. These efforts have brought us close to the goal of a functional unified species-neutral ontology. However, the resulting high-level hierarchy of the ontology lacks the necessary structure to address many uPheno use cases. For example, there is currently not a good set of high-level terms to support ribbon displays at the Alliance of Genome Resources.
This workshop aims to bring together phenotype ontology developers and members of the community with use cases that would benefit from the use of uPheno for phenotype curation.
During the workshop we will:
- Review use cases for uPheno
- Review the current state of uPheno, including:
- the integrated hierarchy
- the patterns and how they are used
- Identify what improvements are needed in uPheno
- Focus on improving the upper-level structure
- Identify other areas that need improvement
- Discuss ways to speed development and increase engagement
Maximizing Community Curation for the Benefit of All
Saturday: 1:30 pm - 5 pm, Organized by Kimberly Van Auken and Daniela Raciti
Biological knowledgebases rely on expert biocuration of the research literature to maintain up-to-date collections of data organized in machine-readable form. However, despite the steady, or even increasing, amount of curatable knowledge, resources supporting expert biocuration are declining, leaving knowledgebases no alternative but to explore additional ways to update and maintain content.
One way in which knowledgebases have addressed this problem is by engaging researchers to help curate their papers, a process generally known as ‘community curation’. As helpful as community curation can be, though, it is not universally adopted and for groups that do have it, there is a wide range of approaches.
In this workshop, we aim to examine the different approaches that groups use, share successes, failures, and ongoing challenges, and produce suggested deliverables for a broader adoption of best practices and tools for effective community curation.
This is a half-day workshop with speaker sessions followed by a panel discussion to produce a list of actionable items to promote community curation.
Speaker Sessions:
- Community curation at non-Model Organism Databases (broader user community)
- Community curation at Model Organism Databases (targeted user community)
- Pre- and post-publishing NLP/AI/ML
- Methods/techniques to serve in community curation
12 speakers will present centered around a focused discussion incorporating topics pertinent to community curation, including:
- Outreach: identifying, training, retaining community curators
- Types of community curated data
- Incentivizing participation
- Integration of AI/ML in community curation workflows
- Sustainability
- Pre- vs post-publication approaches
After the talks, we will have a panel discussion to produce an actionable plan to begin to share resources, expertise, and ideas on community curation with the aim of streamlining and standardizing the author experience and maximizing participation for the benefit of as many knowledgebases as possible.
LANL’s HIV Immunology Database: the first host-pathogen database
Saturday: 2:30 - 5 pm, Organized by Elizabeth-Sharon Fung
(NIH supported)
LA-UR-24-29045
Los Alamos National Laboratory’s HIV Databases (hiv.lanl.gov) consists of two main databases, the HIV Sequence and Immunology Databases, and has been continuously funded by the NIH since 1987. The HIV Sequence Database was founded in 1986 as a repository for the explosion of HIV pathogen sequences, and the HIV Immunology Database, the first host-pathogen database, was added in 1995. This workshop will focus on the HIV Immunology Database. This database contains several sub-databases storing the following information: antibody neutralization activity, antibody contacts and features, epitope variants, and patient information.
In this workshop, we will discuss the contents of the HIV Immunology Database and how they can be used by researchers working on HIV vaccines and treatments. We will also discuss the challenges of curating ever-evolving findings on immune responses to HIV, as well as plans for future improvements to the HIV Immunology Database and its applicability to research on other pathogens.
Information in the HIV Immunology Database is curated from published work. Our database contains records associated with host immune responses to HIV, including cytotoxic and helper T cell epitope responses and host antibodies generated. Our annotation process is highly standardized, and additional quality control checks exist at later time points. Data from our database can be searched, downloaded, or analyzed in web-based tools, and the HIV Immunology Database is connected to the HIV Sequence Database and related databases, allowing users to work with multiple databases in parallel. All of this is dependent upon our database structure, which consists of multiple SQL databases together encompassing hundreds of tables and thousands of relations. In addition, our database hosts many data analysis tools, many of which are applicable to species beyond HIV.
The HIV Immunology Database has significantly contributed to standardizing HIV immunological research. As new research findings emerge, our group must make decisions on what new types of information to curate, and how this information should be stored in our database and presented to users. The challenges of this will be discussed, along with potential updates to our database, including addition of more detailed immunological information, the ability for database users to upload their own data, continual modernization of older database records, and novel manners of presenting large datasets.
Attendees of our workshop should leave with a clear understanding of how our curation model has enabled HIV research and our perspectives on how to stay abreast of evolving findings in host-pathogen research.
Elevating Biological Curators and Knowledgebases in Scholarly Communication
Sunday: 8:30 am - 10:30 am, Organized by Daniela Raciti and Karen Yook
microPublication Biology is a journal dedicated to publishing single-figure findings. A key mission of our journal is to seamlessly incorporate peer-reviewed data into community knowledgebases while reducing the workload on database curators after publication.
In this workshop, we will explore strategies for integrating data curation into the publishing process, with a particular focus on the dual role of managing editors who serve as curators for authoritative databases.
We will introduce an innovative tool designed to identify known biological entities and predict potential ones, streamlining the curation process. This tool not only verifies and extracts bioentities known to the database, but also alerts curators to missing entries that should be added. The tool also assists authors in detecting typographical errors, which can be corrected before publication.
Participants will have the opportunity to interact with microPublication Biology Managing Editors, learn how database curators are crucial to the quality of published research, and sign up for ongoing collaboration or involvement in future data curation initiatives. This workshop is designed for editors, publishers, data curators, and anyone involved in scientific publishing who is interested in improving data quality and curation practices. By sharing our experiences and engaging with curators from diverse knowledgebases, we aim to address the barriers encountered during data curation, reduce time-wasting inefficiencies, and find solutions for these challenges at the pre-publication stage. Our goal is to foster a collaborative environment where curators are central to the publishing process, ensuring data is consistently curated, validated, and optimized for reuse, thereby enhancing the quality and impact of published research.
Data Standardization and Integration with the Bioregistry
Sunday: 8:30 am - 10:30 am, Organized by Charles Tapley Hoyt and Benjamin M. Gyori
The Bioregistry project (https://bioregistry.io, https://github.com/biopragmati..., https://www.nature.com/article...) promotes data integration by cataloging resources that assign persistent identifiers to biomedical concepts.
It supports many linked open data and semantic web users by producing a harmonized and comprehensive prefix map and providing standardized tooling for working with prefixes, uniform resource identifiers (URIs), and compact URIs (CURIEs). The Bioregistry in turn is used by tools like LinkML, data standards like SSSOM, web applications like the EBI Ontology Lookup Service (OLS), and projects like the OBO Foundry and Monarch Initiative (see https://biopragmatics.github.i...).
This two-part workshop will include a lecture and hackathon component.
First, we will give an introduction to the Bioregistry that includes the following:
- An overview of the data model, database, web application, and Python package
- A practical example of using the Bioregistry for data standardization and integration
- Maintenance of the Bioregistry
- Making a new prefix request
- Reviewing a prefix request
- An overview of curation tasks and guides for new contributors (https://biopragmatics.github.io/bioregistry/curation)
Second, we will host a hackathon open to veteran and new contributors. We will work together to address (some of) the following:
- Improve existing records, e.g., by adding contact people
- Resolve open issues for new prefixes and updating existing prefixes (https://github.com/biopragmatics/bioregistry/issues)
- Improve harmonization with other registries
- Integrate Wikidata properties for several domains (taxonomy, bibliometrics, chemistry, etc.)
- Pilot semi-automated new prefix suggestion workflow
- Address use case-specific data standardization and integration scenarios in a "bring your own data" setting
Based on this experience, we expect to write new contribution guidelines and tutorials that will enable additional contributors. The Bioregistry governance model stipulates that all material contributors to the resource are eligible for co-authorship on future papers. If we are able to make substantial contributions during this time, we would also like to write a short conference report and consider outlining an update paper as a follow-up to the original 2022 publication in Nature Scientific Data.
Exploring and Enhancing a Glycan Function Ontology
Sunday: 8:30 am - 12:30 pm, Organized by Michael Tiemeyer
Dynamic changes in protein and lipid glycosylation impact protein homeostasis, membrane functions, development, inflammation, immunity, aging, neural function, pathogen interactions, and almost all aspects of human health from fertilization to death. However, current bioinformatic resources that capture disease and phenotype information focus primarily on the macromolecules represented within the central dogma of molecular biology (DNA, RNA, proteins). In order to gain a more complete understanding of human disease, there is a need to capture the functional impact of glycans and glycosylation on biological processes. We propose a workshop, which would be the third in an ongoing series of workshops sponsored by GlyGen, focused on annotating glycan functions in biological contexts, their digital representation, and integration with existing function annotation efforts.
The first “Workshop on Glycan Function Annotation” was held in conjunction with the 2023 ISB meeting in Padua, Italy and was entitled “Functional Impact of Glycans and their Annotation.” This workshop brought together subject matter experts, tool developers, and biocurators who annotate content that may directly or unknowingly be related to the functional impact of glycans. The attendees identified areas where curators, data wranglers, and text mining experts could collaborate to address gaps in glycan and glycosylation annotations, leverage each other’s work to improve their respective resources, and encourage data sharing amongst resources. Concrete suggestions were developed for improving the visibility and impact of glycan, glycosylation, and glycoconjugate data. A manuscript describing the discussions and conclusions was recently published (https://doi.org/10.1093/database/baae073).
Among the recommendations of the first workshop at ISB2023 was a proposal that the bioinformatics community would benefit greatly from the organization of a second workshop that could harvest input from experts in glycobiology and glycan functions. Thus, a second workshop, entitled “Defining Glycan Functions,” was organized and held in conjunction with the 2023 Society for Glycobiology meeting in Kona, Hawaii. The participants, who together possess deep knowledge of a broad range of glycan functions, developed an elemental foundation for describing vocabularies and concepts capable of capturing glycan functions. The participants acknowledged the need for expanding their framework of functions to include hierarchical relationships between glycan types and specific glycan structures, structural motifs, features, and patterns. A manuscript describing the discussions and conclusions is in preparation.
The participants of the first and second workshops jointly recognized the need to expand discussions across the glycoscience, curation, and informatics community in order to hone annotations into forms that are useful across resources.
The third “Workshop on Glycan Function Annotation,” will be held in conjunction with ISB2025, entitled “Exploring and Enhancing a Glycan Function Ontology,” and will seek the input of participants with expertise in ontologies that describe biological functions. The discussions will build on the outcomes of the first two workshops which identified currently available resources and established an initial ontology for glycan function relationships and associated metadata. It is anticipated that the outcome of the workshop will move the field forward toward finalizing the ontology and its relationships to existing function annotation efforts. This work will lay the foundation for primary data generators and biocurators to collect and organize glycan function annotation as they attempt to enrich our understanding of the impact of glycans and glycosylation across biological domains.
GlyGen is organizing this proposed workshop and expect sthat it will draw interest from ISB2025 attendees who have experience in gene and protein function, ontology developers, as well as curators that focus on the role of post-translational modifications and metabolism in regulating biological activities.
In addition to welcoming any interested ISB meeting attendees, the organizers are also target a group of specific stakeholders to ensure the highest likelihood of generating impactful workshop outcomes.
Biocuration 2025 Hackathon (Part 1)
Sunday: 10:30 am - 12:30 pm, Organized by Charles Tapley Hoyt
Biocuration 2025 Hackathon
Biocurators' interests cover (but are not limited to) the development of standards, annotation of datasets, curation of databases, implementation of software, and deployment of technologies. In many cases, this work is done remotely, collaboratively, and in an open setting. Therefore, we propose a workshop dedicated to hackathons, curate-athons, and other co-working opportunities only possible during the annual international biocuration conference.
While the Biocuration 2025 Hackathon will have space for ad hoc co-working, we have already have planned two tracks (and welcome suggestions for additional tracks):
- OBO Foundry hackathon and curate-athon
- Semantic mappings curate-athon
These tracks have built-in training sessions, meaning that anyone who is interested to learn more or make their first contributions are welcome to join. These sessions' communities follow the open data, open code, open infrastructure (O3) guidelines (https://doi.org/10.1038/s41597-024-03406-w) and also are generous with including contributors as co-authors on manuscripts.
We will use https://bit.ly/biocuration2025-hackathon to solicit additional sessions, distribute additional information about the structure of the planned sessions, and keep notes.
OBO Foundry Hackathon
The Open Biological and Biomedical Ontology (OBO) Foundry (https://obofoundry.org; https://github.com/OBOFoundry/OBOFoundry.github.io; https://doi.org/10.1093/database/baab069) contains some of the most impactful ontologies in the biomedical domain such as the Gene Ontology (GO), Cell Ontology (CL), Disease Ontology (DO), and Uber anatomy Ontology (UBERON). OBO Foundry ontologies commit to curation and maintenance following shared principles (https://obofoundry.org/principles/fp-000-summary.html). In many cases, they use shared tooling. OBO Foundry ontologies together have hundreds of unique contributors, many of whose jobs extend beyond biocuration into specific domain applications. Importantly, many are active members of the ISB who regularly attend its yearly conference.
We propose an open-ended hackathon open to new and veteran OBO Foundry contributors to tackle some of (but not limited to) the following problems:
- Build and maintain the social aspect of the community (spending time together is incredibly valuable!)
- Curation of new terms in various ontologies
- Working through long-standing discussions or issues on various ontologies' issue trackers
- Discussion and improvement to upper-level OBO Foundry ontologies like the Relation Ontology (RO), OBO Metadata Ontology (OMO), Core Ontology for Biology and Biomedicine (COB), and Information Artifact Ontology (IAO)
Conversion of existing ontologies to use the Ontology Development Kit (ODK)
- Improvement of the OBO Foundry website
- Ad-hoc sessions on ontology access, e.g., with the Ontology Access Kit (OAK)
- Discussion and hacking on OBO-adjacent standards, e.g SSSOM for ontology mappings, KGCL for representing change in ontologies, LinkML for data modeling using ontologies
- Improvement of non-OBO Foundry ontologies
Semantic Mappings Curate-athon
Semantic mappings are crucial for data integration efforts that ingest partially overlapping resources. For example, the same disease can appear in the Disease Ontology (DO), Mondo Disease Ontology (MONDO), Medical Subject Headings (MeSH), and dozens of other partially overlapping nomenclature resources.
The Simple Standard for Sharing Ontological Mappings (SSSOM; https://mapping-commons.github.io/sssom; https://doi.org/10.1093/database/baac035) has standardized the way that high quality, precise semantic mappings can be stored, distributed, and processed.
Biomappings (https://github.com/biopragmatics/biomappings) is an open, community-maintained database of semantic mappings that provides a framework for generating predicted mappings and a front-end for curating them. Biomappings uses SSSOM and can be directly incorporated in ontologies via the ODK, such as was done in UBERON (see https://github.com/obophenotype/uberon/pull/3141).
In this short workshop, we will do the following:
- Install and run the Biomappings curation interface
- Make some mapping curations
- Contribute them to the Biomappings databases on GitHub
Given the domain expertise of attendees, we can prepare new predictions appropriate for their ontologies of interest. Otherwise, a few suggested domains are:
- Disciplines and Occupations
- Qualifications and Degrees
- Chemicals
- Pathways and Biological Processes
Genomes Online Database (GOLD) – A curated metadata resource for powering discoveries
Sunday: 10:30 am - 12:30 pm, Organized by TBK Reddy and Supratim Mukherjee
The Genomes Online Database (GOLD) is a metadata management system that catalogs sequencing projects and their associated metadata from around the world. There are three different sources for projects in GOLD: internal projects from the Department of Energy Joint Genome Institute (DOE-JGI), projects entered by external users and those sourced from public databases such as NCBI. The manually curated data in GOLD is organized in a four-level classification system, namely Study, Biosample/Organism, Sequencing Project, and Analysis Project. These four levels contain hundreds of metadata fields including over 100 controlled vocabularies (CVs) containing over 5000 terms, that are presented in an easy-to-use web user interface. This interactive UI facilitates submission of a diverse range of sequencing projects (such as metagenome, metatranscriptome, isolate genome, single-cell genome) and complex analysis projects (such as combined assembly from multiple sequencing projects). It provides a seamless interface with the Integrated Microbial Genomes (IMG) family of analysis tools and supports and the Genomic Standards Consortium (GSC) Minimum Information about any (x) sequence (MIxS) specifications.
This workshop will feature a tutorial on how to enter sequencing projects in GOLD, exploring GOLD, and examples of metadata driven discoveries. Participants will learn about our efforts to follow FAIR data principles by integrating MIxS environmental packages, Research Organization Registry (ROR) and The Genome Taxonomy Database (GTDB), to name a few. They will get familiarized with custom searches by using a wide range of predefined metadata filters. Participants will have the opportunity to perform their own search using free text fields and metadata terms. We will go over select use case scenarios where researchers used GOLD’s metadata for discoveries and hypotheses testing to demonstrate how curated metadata can be used to power discoveries. Participants will also learn how to explore NCBI SRA data within GOLD using NCBI identifiers (such as SRA IDs, Taxonomy Name, Sequencing Strategy and more).
This workshop will also introduce GOLD’s canonical naming system for microbiome samples. Contrary to isolate genomes that have standardized names governed by established principles of phylogeny, microbiomes lack systematic naming standards. As a result, many microbiomes in public repositories have cryptic, esoteric names. To address this shortcoming, GOLD has developed a methodology to use distinct metadata features of a sample to construct a standardized name. Participants will learn how to use a combination of metadata fields like habitat, community, location, and distinct identifier to construct a microbiome project name that captures the entire contextual information of a sample.
AI and Biodata Resources: Implications for Sustainability and Best Practices in Biocuration (Part 1)
Sunday: 10:30 am - 12:30 pm
Organizers: Valerio Arnaboldi (WormBase, Alliance of Genome Resources), Guy Cochrane (Global Biodata Coalition), Chuck Cook (Global Biodata Coalition), Matt Jeffryes (Europe PMC), Melissa Harrison (Europe PMC), Lynn M. Schriml (Human Disease Ontology), and Pengyuan Li (IBM, Alliance of Genome Resources)
Workshop premise: The integration of Machine Learning (ML) and Artificial Intelligence (AI) into workflows of biodata resources represents a paradigm shift in how biological data is extracted, curated, and shared. The recent advances and proliferation of Large Language Models (LLMs) are accelerating this transformation, bringing both opportunities and challenges. LLMs, while powerful, are often thought of as being capable of solving any problem, leading to a disconnect with their fit-for-purpose. There is a pressing need for a shared understanding of what these tools can and cannot realistically deliver, and how these capabilities enhance, or detract from, human curation and management of biodata resources. In particular, will these new tools improve efficiency, leading to more sustainability, or will the costs exceed gains in efficiency?
This workshop aims to map the current landscape of ML and AI applications in biocuration, identify available tools, and discuss the successes and challenges faced while incorporating these technologies into biodata resources, with particular emphasis on sustainability, cost, and transparency of their usage. The workshop will serve as a platform for discussing how to make ML more accessible and beneficial for biodata resources and the biocuration community at large, and how to bridge the gap between computer science and biocuration, improving standards for sharing FAIR datasets that can attract more ML and AI experts to the biocuration field. The workshop will foster a collaborative environment where biocurators, biodata resources representatives, and ML experts can share their experiences, learn about new tools, and contribute to the development of ML solutions and integrate them into biodata resources.
Key sustainability and impact-related topics to be addressed may include: mapping of curation workflows and identifying the points at which ML could be introduced, monitoring on deployed tools and track their development in comparison to previous editions of this workshop at ISB meetings, sustainability and costs of using ML solutions in biodata resources, identifying what biocurator needs may be met by ML, benchmarking and validation of ML to determine which technologies may be truly beneficial to biocuration. Talks may also explore the increasing role of biocurators in the production of gold standard datasets necessary for training of ML models, metadata standardization in order to increase the utility of ML augmented data, transparency in the use of ML to ensure users and communities are aware of where and when automated curation has been deployed, species bias in the biomedical literature and the likely impact of this on ML models trained from it.
The workshop would use a number of formats to encourage participation from all attendees, both in person and remote, including a pre-workshop survey to gather community members’ experience with ML, online whiteboard based collaboration to annotate and visualize curation workflows, panel discussion involving biocurators, representatives of biodata resources, ML experts and industry representatives to discuss the pros and cons of ML deployment in biocuration, and short talks giving insight into specific deployments of ML and lessons learned, and highlighting innovative solutions to common problems. The workshop organizers who represent three GBC Global Core Biodata Resources: the Alliance of Genome Resources, the Human Disease Ontology, and Europe PMC, will lead through presentation of their own experiences.
As an outcome from the workshop, we aim to develop concrete action items for the community to take around the subjects discussed, establishing community channels and fostering collaboration between biocurators and ML tool developers through the Global Biodata Coalition, and a roadmap for ML utilization in biocuration giving guidance on practical applications and transparency on the use of ML in biocuration tasks.
Biocuration 2025 Hackathon (Part 2)
Sunday: 1:30 pm - 5 pm, Organized by Charles Tapley Hoyt
Biocuration 2025 Hackathon
Biocurators' interests cover (but are not limited to) the development of standards, annotation of datasets, curation of databases, implementation of software, and deployment of technologies. In many cases, this work is done remotely, collaboratively, and in an open setting. Therefore, we propose a workshop dedicated to hackathons, curate-athons, and other co-working opportunities only possible during the annual international biocuration conference.
While the Biocuration 2025 Hackathon will have space for ad hoc co-working, we have already have planned two tracks (and welcome suggestions for additional tracks):
- OBO Foundry hackathon and curate-athon
- Semantic mappings curate-athon
These tracks have built-in training sessions, meaning that anyone who is interested to learn more or make their first contributions are welcome to join. These sessions' communities follow the open data, open code, open infrastructure (O3) guidelines (https://doi.org/10.1038/s41597-024-03406-w) and also are generous with including contributors as co-authors on manuscripts.
We will use https://bit.ly/biocuration2025-hackathon to solicit additional sessions, distribute additional information about the structure of the planned sessions, and keep notes.
OBO Foundry Hackathon
The Open Biological and Biomedical Ontology (OBO) Foundry (https://obofoundry.org; https://github.com/OBOFoundry/OBOFoundry.github.io; https://doi.org/10.1093/database/baab069) contains some of the most impactful ontologies in the biomedical domain such as the Gene Ontology (GO), Cell Ontology (CL), Disease Ontology (DO), and Uber anatomy Ontology (UBERON). OBO Foundry ontologies commit to curation and maintenance following shared principles (https://obofoundry.org/principles/fp-000-summary.html). In many cases, they use shared tooling. OBO Foundry ontologies together have hundreds of unique contributors, many of whose jobs extend beyond biocuration into specific domain applications. Importantly, many are active members of the ISB who regularly attend its yearly conference.
We propose an open-ended hackathon open to new and veteran OBO Foundry contributors to tackle some of (but not limited to) the following problems:
- Build and maintain the social aspect of the community (spending time together is incredibly valuable!)
- Curation of new terms in various ontologies
- Working through long-standing discussions or issues on various ontologies' issue trackers
- Discussion and improvement to upper-level OBO Foundry ontologies like the Relation Ontology (RO), OBO Metadata Ontology (OMO), Core Ontology for Biology and Biomedicine (COB), and Information Artifact Ontology (IAO)
Conversion of existing ontologies to use the Ontology Development Kit (ODK)
- Improvement of the OBO Foundry website
- Ad-hoc sessions on ontology access, e.g., with the Ontology Access Kit (OAK)
- Discussion and hacking on OBO-adjacent standards, e.g SSSOM for ontology mappings, KGCL for representing change in ontologies, LinkML for data modeling using ontologies
- Improvement of non-OBO Foundry ontologies
Semantic Mappings Curate-athon
Semantic mappings are crucial for data integration efforts that ingest partially overlapping resources. For example, the same disease can appear in the Disease Ontology (DO), Mondo Disease Ontology (MONDO), Medical Subject Headings (MeSH), and dozens of other partially overlapping nomenclature resources.
The Simple Standard for Sharing Ontological Mappings (SSSOM; https://mapping-commons.github.io/sssom; https://doi.org/10.1093/database/baac035) has standardized the way that high quality, precise semantic mappings can be stored, distributed, and processed.
Biomappings (https://github.com/biopragmatics/biomappings) is an open, community-maintained database of semantic mappings that provides a framework for generating predicted mappings and a front-end for curating them. Biomappings uses SSSOM and can be directly incorporated in ontologies via the ODK, such as was done in UBERON (see https://github.com/obophenotype/uberon/pull/3141).
In this short workshop, we will do the following:
- Install and run the Biomappings curation interface
- Make some mapping curations
- Contribute them to the Biomappings databases on GitHub
Given the domain expertise of attendees, we can prepare new predictions appropriate for their ontologies of interest. Otherwise, a few suggested domains are:
- Disciplines and Occupations
- Qualifications and Degrees
- Chemicals
- Pathways and Biological Processes
scFAIR: FAIRification of single-cell data
Sunday: 1:30 - 5 pm, Organized by Frederic Bastian and Marc Robinson-Rechavi
Single-cell functional genomics is a major field of biology, which is bringing essential insight into the life sciences. Single-cell data are rapidly increasing both in quantity and in diversity, but lack method and metadata standardization. While some large projects have clear standards of reporting, most datasets in biological databases have partial or non standardized metadata. This leads to multiple non-compatible standards across datasets, and limits reusability, which in turn presents challenges to make these data useful to an increasing community of specialists and non-specialists. There is a need for standards in the way single-cell data are stored and annotated, especially for cell type and other associated information. Indeed, metadata is critical to the capacity to use these large and potentially very informative datasets. It includes protocols, which constrain which transcripts were accessible or which normalizations are relevant, the association between barcodes and annotations, or the methods used to identify cell types. Existing ontologies and controlled vocabularies are not used systematically, even when information is reported.
To address these limitations, the scFAIR consortium has the mission to build a collaborative platform supporting and disseminating ORD practices for the single-cell genomics community, both for sharing datasets and their metadata, and for standardizing the way data are shared across datasets. scFAIR includes members from several international organizations, and already provides tools to query datasets across resources, or standards to share essential information, such as marker genes used to identify cell types. scFAIR work is accessible at https://sc-fair.org/ .
During this workshop, participants will have the opportunity to shape the definition of this collaborative platform, by sharing their needs and best practices. They will also receive all necessary information to enroll in the consortium, or to use the tools developed by the consortium. This workshop will notably cover:
- how to describe and capture information about processing pipelines, in order to reproduce analyses, and determine how cell features were identified
- how to provide in a standard way information about barcodes identifying cells, and the raw data of the analysis
- how to annotate cell clusters with controlled vocabularies and ontologies
- best practices in quality control
- best practices in cell type inference.
On each topic, after an introduction by a field expert, we will ask participants to share the difficulties they face in their own work, and what they miss to leverage to their full potential existing single-cell data. We will then work together to synthesize recommendations. We expect that, as a result, participants will acquire an improved knowledge of the existing landscape in single-cell biocuration, and a collaborative involvement in the future of Open Research Data in scFAIR.
AI and Biodata Resources: Implications for Sustainability and Best Practices in Biocuration (Part 2)
Sunday: 1:30 - 5 pm
Organizers: Valerio Arnaboldi (WormBase, Alliance of Genome Resources), Guy Cochrane (Global Biodata Coalition), Chuck Cook (Global Biodata Coalition), Matt Jeffryes (Europe PMC), Melissa Harrison (Europe PMC), Lynn M. Schriml (Human Disease Ontology), and Pengyuan Li (IBM, Alliance of Genome Resources)
Workshop premise: The integration of Machine Learning (ML) and Artificial Intelligence (AI) into workflows of biodata resources represents a paradigm shift in how biological data is extracted, curated, and shared. The recent advances and proliferation of Large Language Models (LLMs) are accelerating this transformation, bringing both opportunities and challenges. LLMs, while powerful, are often thought of as being capable of solving any problem, leading to a disconnect with their fit-for-purpose. There is a pressing need for a shared understanding of what these tools can and cannot realistically deliver, and how these capabilities enhance, or detract from, human curation and management of biodata resources. In particular, will these new tools improve efficiency, leading to more sustainability, or will the costs exceed gains in efficiency?
This workshop aims to map the current landscape of ML and AI applications in biocuration, identify available tools, and discuss the successes and challenges faced while incorporating these technologies into biodata resources, with particular emphasis on sustainability, cost, and transparency of their usage. The workshop will serve as a platform for discussing how to make ML more accessible and beneficial for biodata resources and the biocuration community at large, and how to bridge the gap between computer science and biocuration, improving standards for sharing FAIR datasets that can attract more ML and AI experts to the biocuration field. The workshop will foster a collaborative environment where biocurators, biodata resources representatives, and ML experts can share their experiences, learn about new tools, and contribute to the development of ML solutions and integrate them into biodata resources.
Key sustainability and impact-related topics to be addressed may include: mapping of curation workflows and identifying the points at which ML could be introduced, monitoring on deployed tools and track their development in comparison to previous editions of this workshop at ISB meetings, sustainability and costs of using ML solutions in biodata resources, identifying what biocurator needs may be met by ML, benchmarking and validation of ML to determine which technologies may be truly beneficial to biocuration. Talks may also explore the increasing role of biocurators in the production of gold standard datasets necessary for training of ML models, metadata standardization in order to increase the utility of ML augmented data, transparency in the use of ML to ensure users and communities are aware of where and when automated curation has been deployed, species bias in the biomedical literature and the likely impact of this on ML models trained from it.
The workshop would use a number of formats to encourage participation from all attendees, both in person and remote, including a pre-workshop survey to gather community members’ experience with ML, online whiteboard based collaboration to annotate and visualize curation workflows, panel discussion involving biocurators, representatives of biodata resources, ML experts and industry representatives to discuss the pros and cons of ML deployment in biocuration, and short talks giving insight into specific deployments of ML and lessons learned, and highlighting innovative solutions to common problems. The workshop organizers who represent three GBC Global Core Biodata Resources: the Alliance of Genome Resources, the Human Disease Ontology, and Europe PMC, will lead through presentation of their own experiences.
As an outcome from the workshop, we aim to develop concrete action items for the community to take around the subjects discussed, establishing community channels and fostering collaboration between biocurators and ML tool developers through the Global Biodata Coalition, and a roadmap for ML utilization in biocuration giving guidance on practical applications and transparency on the use of ML in biocuration tasks.
Biocuration Career Opportunities
Wednesday: 1:00 pm - 2:00 pm, Organized by Nicole Vasilevsky and Randi Vita
The International Society for Biocuration (ISB) is dedicated to advancing the field of biocuration and supporting the professional growth of curators. As a relatively under-recognized field, biocuration faces challenges in attracting new talent and defining career advancement pathways. This year's careers workshop aims to highlight current career opportunities within biocuration and attract new trainees to the field.
The workshop will target students, postdocs, and other academic trainees and staff in the Kansas City area, in addition to the Biocuration conference attendees.
We will invite people in the field who have current job openings or anticipate hiring soon. These speakers will discuss available opportunities and various roles within the field in a relaxed setting.
Our goal for the workshop is to reach a broader audience, raise awareness about biocuration, and provide existing curators with valuable career opportunities. Additionally, it will offer hiring managers a platform to promote their available positions and institutional opportunities.
Schedule of events
Saturday, April 5
9:00-12:30 PM - Morning workshops
1:30-5:00 PM - Afternoon workshops
Sunday, April 6
8:30-12:30 PM - Morning workshops
1:30-5:00 PM - Afternoon workshops
Monday, April 7
9:00-12:00 PM – Plenary session
1:00-5:00 PM – Plenary session
5:00-6:30 PM – Poster session 1
Tuesday, April 8
9:00-12:00 PM – Plenary session
1:00-4:30 PM – Plenary session
4:30-6:00 PM – Poster session 2
7:00-10:00 PM – Social dinner
Wednesday, April 9
9:00-12:00 PM – Plenary sessions
1:00-2:00 PM – Biocuration Career Opportunities Workshop
2:00-3:00 PM – Closing
Conference Venue
Stowers Institute
Our 10-acre campus is situated in the heart of Kansas City. Located next to the University of Missouri-Kansas City and The Nelson-Atkins Museum of Art, our members are part of an academic neighborhood. You can immerse yourself in the arts, visit landmarks such as the Linda Hall Scientific Research Library, and find inspiration to fuel creative thinking.
At the Stowers Institute you are also within walking distance to Loose Park, the Trolley Trail, the Brookside Neighborhood, and the Country Club Plaza. You have convenient access to some of the best coffee shops, restaurants, and shopping the city has to offer.
Kansas City, Missouri
Kansas City may be located in the heart of the country, but it’s certainly not flyover country. Known fondly as the “City of Fountains” and “The Paris of the Plains,” you’ll fall in love with Kansas City’s unique architecture, stunning beauty, and dynamic culture. Thanks to explosive growth in the past decade and a strong sense of Midwest pride, more than 2 million people call Greater Kansas City home, making it one of the top 30 most populated metros in the U.S., situated conveniently between both coasts.
Lodging
Hotel information:
Aloft Kansas City Country Club Plaza is the preferred conference hotel. Morning and evening shuttles will be offered between the Aloft Hotel and Stowers Institute. For those who prefer to walk, plan for a 1.3 mile urban walk to Stowers Institute.
There are several other nearby hotels within 1.5 miles of Stowers Institute, including Cascade Hotel Kansas City, which is next door to the Aloft Hotel for convenient shuttle access, as well as Hilton Kansas City Country Club Plaza
Shuttle information:
Complimentary shuttle service will be offered each morning and evening on full conference days between the Aloft Kansas City Country Club Plaza and Stowers Institute. Neighboring Cascade Hotel Kansas City sits next door and is only steps away from the Aloft for convenient shuttle access.
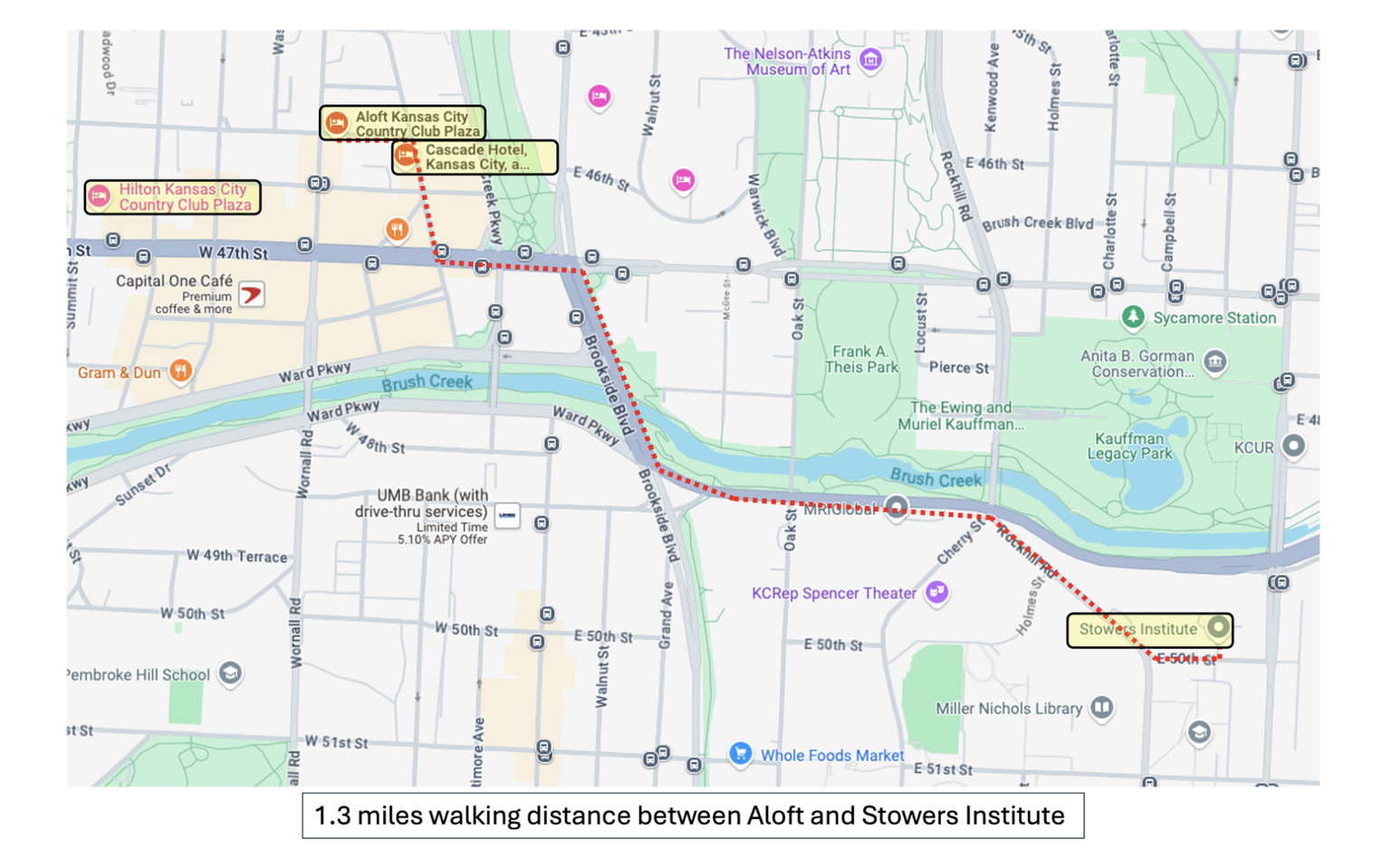
View map details, walking path, and and location of events here.
Essential Meeting Information
Location: Stowers Institute for Medical Research – 1000 E. 50th Street, Kansas City, MO 64110 USA
For turn-by-turn directions to the Institute from where you are, click here.
Arrival: Check-in & poster set up will start on Monday, April 7 at 8:00 am. Opening remarks and announcements will begin promptly at 9:00 am.
Nearest Airport: Kansas City International Airport: https://www.flykci.com/
Ground Transportation: There are a number of app-based ride share services, including Lyft, Uber and zTrip, as well as shuttle services offering transportation to and from Kansas City International Airport.
Lodging: Aloft Kansas City Country Club Plaza is the preferred conference hotel. Morning and evening shuttles will be offered between the Aloft hotel and the conference venue.
There are several other nearby hotels within 1.5 miles of Stowers Institute, including Cascade Hotel Kansas City, which is next door to the Aloft Hotel for convenient shuttle service, as well as Hilton Kansas City Country Club Plaza.
Conference Shuttles: Complimentary shuttle service will be offered on full conference days for one hour in the morning and one hour in the evening between the Aloft Kansas City Country Club Plaza and the Stowers Institute for Medical Research. In addition, a shuttle will run to and from the social dinner.
Parking: Visitor parking is available in the parking structure located between the Administration Building (to your right as you enter the campus) and the Research Building (to your left). Please park on the 5th floor of the parking garage and take the elevator down to the 1st floor to enter the Research Building.
Abstract Submission: Everyone is welcome to submit an abstract. On the submission form, you may choose to be considered for a short talk or a poster.
Abstract Deadline: November 14, 2024
Poster Dimensions: All posters should be horizontal in orientation.
Maximum poster dimensions: Width – 6 ft. (182 cm), Height – 4 ft. (121 cm)
Shipping and Printing: Local FedEx Office – 554 Westport Rd, Kansas City, MO 64111 US https://local.fedex.com/mo/kansas-city/office-0334/
International Attendees: Send requests for Visa letters to biocuration2025@stowers.org. Registration is required prior to issuing the Visa letter.
Departure: The meeting will conclude on Wednesday, April 9, 2025 at 3:00 PM
Cancelation Policy: Cancelations must be notified in writing to biocuration2025@stowers.org. Refunds will be made under the following conditions:
- For cancelations until December 31, 2024: full refund of registration fees received
- For cancelations between January 1-March 1, 2025: refund of 70% of registration fees received
- For cancelations after March 1, 2025: no refund will be possible
Questions? Don’t hesitate to reach out to biocuration2025@stowers.org
Stowers Institute
Campus map

Learn more about the Stowers Institute
Exploring the fundamental processes of biology is essential in our quest to understand the secrets of life. Stowers Institute research focuses on foundational research in organisms, tissues, and cells to understand the many mechanisms underlying human health and disease.






