In The News

07 January 2026
Investigator Kamena Kostova, named ‘Cell Scientist to Watch’
From the Journal of Cell Science, Investigator Kamena Kostova named a 'Cell Scientist to Watch'
Read Article
By Marla Vacek Broadfoot
The average American adult has an IQ of 98, earns an annual income of $46,550, checks their phone 80 times a day, watches television 35 hours a week, eats 193 pounds of meat a year, and lives until the age of 78.
These statistics might give a snapshot of life in the United States, but in reality no one is average, at least not by all parameters. Reports that lump all 323 million Americans together and look at their average aptitudes, behaviors, and life trajectories inevitably obscure what makes each person unique. They lose sight of the many geniuses, billionaires, bibliophiles, technophobes, vegetarians, and octogenarians who call the United States home. Looking more closely at how individuals differ from statistical averages, as well as how they differ from other individuals, could reveal particular attributes that influence their diverse activities and roles in society.
The same can be said for research on the inner workings of the human body. For decades, researchers have used traditional biology tools to study entire populations of cells—stem cells, hair cells, and cancer cells, just to name a few. These efforts have generated important insights into the mechanisms that underlie health and disease, but they are still predicated on the average life of the average cell. Many scientists at the Stowers Institute and other research institutions now believe if they want to know what makes each cell tick—and to pinpoint those with the extraordinary ability to fuel the spread of cancer or repair damaged tissues—they need to do it at the single-cell level.
“Looking at the single-cell level gives you a lot more power to understand in detail how processes occur in individual cells—which tells you something totally different, and I think much more valuable, than looking at an aggregated pooled sample,” says Andrew Box, senior laboratory manager of the Stowers Cytometry Facility.
Scientists have long suspected that not all cells were created equal, but until recently they lacked the tools to fully explore the role single cells play in biological phenomena like cancer and regeneration. “The field of single-cell analysis has been propelled in recent years by technological advances in laser capture microscopy and microfluidics that enable researchers to isolate and analyze single cells from tissue sections or bulk cell populations, respectively,” says Paul Kulesa, PhD, director of the Stowers Imaging Center and one of the first adopters of single-cell analysis at the Institute.
About five years ago, advances in -omics technology—designed to detect genes (genomics), transcripts (transcriptomics), proteins (proteomics), and metabolites (metabolomics)—made it possible for researchers to gain a glimpse into the complicated internal dialogue taking place in a single cell. They were surprised to find that cells that appeared to be of the same “type” were even more heterogeneous than they once thought. Previous estimates put the number of cell types in the human body at 200. Scientists now think we could house an order of magnitude more.
The introduction of automation, advances in chemical reagents, and miniaturization of reactions have made single-cell analysis techniques more mainstream, and as a result the pace of discovery has accelerated, says Anoja Perera, senior laboratory manager of the Molecular Biology Facility at Stowers. “It’s amazing to think how far we have come,” Perera says. “The single-cell field has grown so fast. Now, we can study the gene expression of thousands of individual cells, in just a few days, for a fraction of the cost.”
How it works
So how do Stowers scientists study cells at the single-cell level? Their exact methods vary depending on what cell or model organism they are studying, and what question they are trying to answer. However, most scientists follow the same basic recipe, involving three to four steps. First, the scientists break up the collections of cells that make up their biological sample—perhaps a piece of tissue from a zebrafish or a colorectal tumor from a mouse—into single cells. This step sounds relatively simple, but it is often the most onerous. As soon as the cells are removed from an organism, their behavior starts to change. Their gene expression patterns shift as they go into a kind of cellular shock and begin to die off. At Stowers, specialists in the cytometry and molecular biology groups work with researchers to help them hone their methods of preparing samples—which could involve separating cells with chemicals or by mechanical means—to save as many cells as possible.
Jason Morrison, a research specialist in the Kulesa Lab, has been using single-cell analysis in chick embryos to look for a unique molecular signature shared by the most invasive neural crest cells that travel long distances from the brainstem to build tissues elsewhere in the body. Together with functional testing to confirm their results, this information would shed light on the genes critical to embryonic cell invasion and allow comparison to other molecular signatures being identified in metastatic cancer, wound healing, and the immune response. He says he has several collaborators at other institutions who covet his setup. “We want to capture the science before the cells realize they are no longer where they are supposed to be,” Morrison says. “At Stowers, this is made seamless through the close coordination of people and different core facilities all in the same building.”
After dissociating their tissues into single cells, researchers might take their samples to the Cytometry Facility if they need to further sort out specific cells they want to study. If they were studying the regeneration of zebrafish sensory organs, for instance, they might use a marker — an antibody or a dye—to label a certain type of stem cell. Then a flow cytometer could detect the marker on those cells, collect them in a tube, and discard the rest. Box says their newest cell analyzer can look for 24 different colors, meaning they could, in theory, stain for different proteins on the surface of cells, different levels of DNA or RNA inside the cells, and different subcellular compartments like mitochondria or endoplasmic reticulum, all at the same time. He and Stowers Investigator Linheng Li, PhD, have discussed using this 20-plus color approach to isolate the many different cell types present in intestinal and colon tumors in his mouse models of cancer.
Once the scientists have isolated and separated their cells, they measure the expression of whatever interests them — perhaps RNA transcripts or protein — in each. This is where the advances in technology really shine. In January 2017, the Institute acquired a cutting-edge instrument called the 10x Chromium, a shoebox-sized machine that can partition cells into individual droplets, alongside all the reagents needed to conduct single-cell transcriptomics. Each individual cell is tagged with its own molecular barcode, and each of the tens of thousands of RNA transcripts receive a unique barcode as well. After all the transcripts are sequenced, scientists can use these barcodes to figure out which genes are active in each cell. For example, Stowers Investigator Ting Xie, PhD, has used the technique to investigate how the niche that serves as a home base for stem cells orchestrates their differentiation into different cell types in ovarian tissues of the fruit fly. “The single-cell sequencing technology offers a unique opportunity for addressing this important question, which was almost impossible in the past,” Xie says.
Finally, having gathered all the data, researchers set about analyzing their results. Whereas steps one through three could take days, this step could take months, even years. “The amount of data that comes out of each of these experiments is enormous—looking at the levels of thousands of different RNA or protein molecules in individual cells, a thousand cells at a time, and comparing changes in expression as the embryo grows,” says Kulesa, who has used single-cell analysis to record the ups and downs of gene expression in single neural crest cells during different stages of migration. “The bioinformaticians have to develop new ways of coordinating, interrogating, and helping to interpret all of that information.”
Stowers Investigator Tatjana Piotrowski, PhD, agrees the data analysis involved in single-cell studies can be cumbersome and requires a certain level of expertise. Her lab has been using single-cell analysis to look for genes that are turned on during the regeneration of hair cells in zebrafish, in the hopes of identifying molecular targets to treat the inner ear defects that cause hearing loss in humans. She has found that the data generated is often fraught with noise. “You need a bioinformatician to parse through the data,” she says. “I couldn’t do anything with it on my own.” That’s why Piotrowski decided about a year ago to add a bioinformatician, Daniel Diaz, to her team.
Bioinformaticians like Diaz collaborate with fellow lab members to develop testable models from these complex datasets. It is an iterative process, which eventually takes the researchers back to where they began—the bench. “The biology is the endpoint, and most importantly, it’s the part you always need to keep in mind during an analysis,” Diaz says. “So, in order to know whether or not a particular model is optimal, you as an analyst either have to have a tight collaboration with the research lab, or if you work in a lab, have a firm grasp of the system you are studying.”
More cells, more parameters
With a new model in hand, Piotrowski and other Stowers scientists can begin to manipulate genes in single cells to test whether the patterns uncovered by their sophisticated data analyses hold true in real life. In the end, each single-cell experiment drives the next experiment, and the next. “The process took over a year, where we sat down around a table with members of the cytometry, molecular biology, and bioinformatics teams, and tried to figure out how to do this project,” Piotrowski says. “We had meetings after we did experiments to discuss results and what could be improved. I could not have done these experiments at any other place, because the scientific support groups here were so fantastic in working together and pulling us in the same direction.”
The collaborative environment at the Institute means no one researcher needs to be an expert on everything, an asset in a rapidly evolving field like single-cell analysis. As little as five years ago, scientists would design an entire experiment around a handful of single cells. Today, the 10x Chromium can analyze 80,000 at a time. Some scientists are eager to design experiments that analyze even more cells—hundreds of thousands, even millions—to generate even cleaner, less noisy data. Looking at more cells not only gives their experiments more statistical power, but it also increases the chances that scientists will capture cells in as many states as possible, so they don’t miss important biological states that could drive health and disease.
Stowers researchers are also adopting novel technologies that will enable them to glean more information from each and every cell passing through their hands. Jennifer Kasemeier, PhD, a senior research specialist in the Kulesa Lab, has been working with a powerful new instrument called Milo that can measure the expression of multiple proteins in each of thousands of single cells in a single run. She sees the future of single-cell research as one in which scientists can look at RNA transcripts, proteins, and a number of other markers simultaneously, to generate a more comprehensive picture of cellular activity so they can spot the handful of cells predicted to become metastatic.
“In cancer cell invasion, the whole tumor is not moving to a new site in the body—it is being led by a single cell, or a few cells. We want to figure out what differentiates those cells from the others,” Kasemeier says. She recently demonstrated that a signal called nerve growth factor, or NGF, could reprogram metastatic melanoma cells into a more benign cell type. “If we could identify and target bad cells before they invade other tissues, it could have tremendous power therapeutically.”
Ultimately, the goal of all single-cell analyses—whether they are on colon tumors in mice, stem cells in fruit flies, regeneration in zebrafish, or cancer metastasis in chicks—is to see what couldn’t be seen before. To look beyond the average behavior of an average cell to gain a greater understanding of what makes each cell unique, recognizing that with every new insight comes an opportunity to dig deeper, ask bigger questions, and fundamentally change our view of the world.
In The News

07 January 2026
From the Journal of Cell Science, Investigator Kamena Kostova named a 'Cell Scientist to Watch'
Read Article
#Stowers25: Celebrating 25 Years
06 January 2026
Alejandro Sánchez Alvarado, Ph.D., reflects on a year of discovery, gratitude, and the community that helps support our mission.
Read Article
In The News
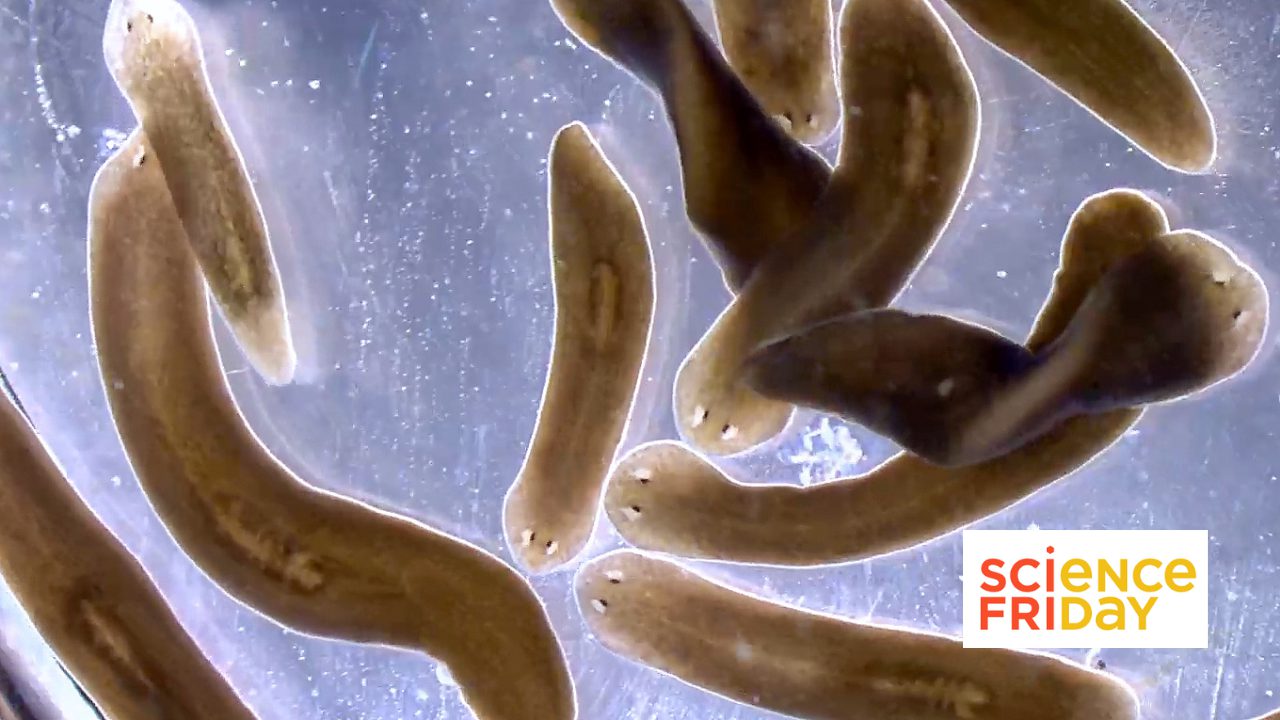
01 January 2026
From Science Friday, President and CSO Alejandro Sánchez Alvarado talks about the science of regeneration and the biology lessons we can carry into the new year.
Read Article
