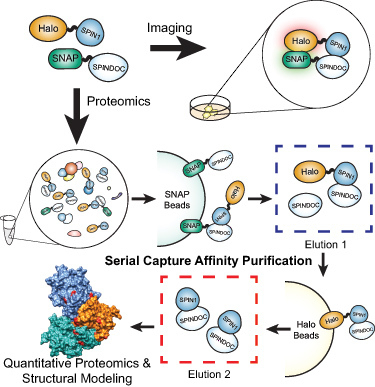
A report published online November 30, 2020, in the Proceedings of the National Academy of Sciences of the USA describes the approach, which includes a double purification technique, Serial Capture Affinity Purification (SCAP), to increase the quality of the protein complex to be analyzed, a frequent hurdle when determining protein structures.
“Structure and function are closely connected in many aspects of biology,” says Xingyu Liu, a predoctoral researcher in the Graduate School of the Stowers Institute and first author of the article. “A protein complex must be assembled in a certain way, with a particular shape in space, in order to carry out its normal function.”
“In many cases, researchers have identified proteins that make up a protein complex,” Liu says. “The individual structures of some of those proteins are known,” Liu says. “But much less is known about exactly how the proteins come together and form the larger assembly.”
By figuring out the 3D structures of protein complexes, researchers can uncover details about how the proteins interact with each other and how these interactions may impact specific functional regions, or domains, of each protein. This type of information can help researchers understand how protein complexes function normally and how certain mutations or cellular states might alter protein complex structure and function, potentially leading to unhealthy conditions or disease.
“We decided to apply our method to a protein complex containing Spinlin1 (SPIN1) and SPINDOC proteins. Spinlin1 is a chromatin reader protein involved in cell signaling and has been implicated in human cancer,” says Liu. “A key feature of the experiment was attaching different affinity tags to Spindlin1 and SPINDOC proteins so that we could perform multi-step affinity purification.”
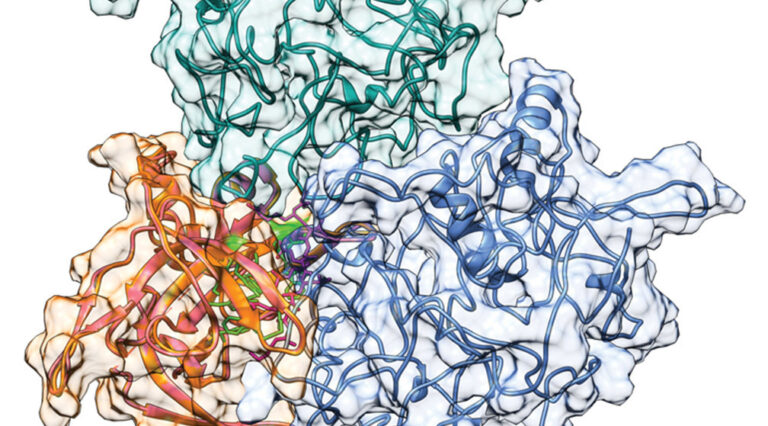
The researchers combined serial capture affinity purification (SCAP), which reduces the complexity and increases the purity of protein complexes, with crosslinking mass spectrometry, molecular modeling, and protein docking technologies. They generated a heterotrimer model consisting of one Spindlin1 protein and two SPINDOC proteins. The protein interactions were validated using live imaging techniques in human tissue culture cells which provided evidence for direct interaction and co-diffusion of the proteins.
“We were excited to test our integrative method and determine a 3D model for the subassembly of three component proteins of the SPIN1-SPINDOC complex,” says Liu. The approach was developed as a generic pipeline with the potential to be applied broadly to protein complexes in tissue culture cells to figure out the structural and functional significance of protein interactions and how they fit into larger molecular networks inside of cells.
The approach also allows researchers to assess interactions between component proteins in vivo using live cell imaging techniques. From there, a larger concept of ProteoCellomics was defined by coauthor and study lead Michael Washburn, PhD, where both ex vivo and in vivo characterizations of protein-protein interactions are combined.
Other coauthors of the study included Ying Zhang, PhD, Zhihui Wen, Yan Hao, Charles A.S. Banks, PhD, Jeffrey J. Lange, PhD, Brian D. Slaughter, PhD, Jay R. Unruh, PhD, Laurence Florens, PhD, Susan M. Abmayr, PhD, and Jerry Workman, PhD.
This work was supported by the Stowers Institute for Medical Research and the National Institute of General Medical Sciences of the National Institutes of Health (awards R35GM118068 to JLW and RO1GM112639 to MPW). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
About the Stowers Institute for Medical Research
The Stowers Institute for Medical Research is a non-profit, basic biomedical research organization dedicated to basic research – the critical first step in the quest for new medical diagnostics, therapies and treatments. Jim Stowers, founder of American Century Investments, and his wife, Virginia, opened the Institute in 2000. Since then, the Institute has spent over one billion dollars in pursuit of its mission.
Currently, the Institute is home to about 500 researchers and support personnel, over 20 independent research programs, and more than a dozen technology development and core facilities. Learn more about the Institute at www.stowers.org and about its graduate program at www.stowers.org/gradschool.