In The News

07 January 2026
Investigator Kamena Kostova, named ‘Cell Scientist to Watch’
From the Journal of Cell Science, Investigator Kamena Kostova named a 'Cell Scientist to Watch'
Read Article
News
“It may not matter exactly where a regulatory element is on a chromosome, but how it interacts with target genes in time and space to build a body is really important.”

Mouse head-to-tail body plan is achieved through the dynamic coordination and regulation of a cluster of Hox genes by three shared enhancers. Cover image designed by Zainab Afzal, Ph.D.
By Rachel Scanza, Ph.D.
The bending and spiraling of light due to dark matter’s gravitational pull during star and galaxy formation delivers exquisitely detailed displays of the early universe, courtesy of the James Webb Space Telescope. In astronomy, distance is not the appropriate metric to measure the age of cosmic entities like stars—after all, the universe is expanding. Rather, it’s time. Time since the big bang.
Moving from macro to micro—from the universe to genes—new research from the lab of Investigator Robb Krumlauf, Ph.D., at the Stowers Institute for Medical Research expands our understanding of how regulatory elements situated among their target genes on a linear strand of chromosomal DNA actually exert their effects in a manner more akin to relativity. Chromosome shape is dynamic and can bend and loop, like gravity-drawn light, to bring genes and their regulators together in time.
“It may not matter exactly where a regulatory element is on a chromosome, but how it interacts with target genes in time and space to build a body is really important,” said Krumlauf. Understanding gene regulation dynamics and body patterning mechanisms tells us more about how the features of the body are regulated in development, disease, and evolution.
Hox genes are evolutionarily conserved, master regulators of an organism’s body plan—from sea anemones to mice to humans. And they are arranged in a linear sequence on a chromosome that determines the order of developing body segments. However, regulatory sequences of DNA called enhancers that precisely control expression of these genes are frequently far away from their targets.
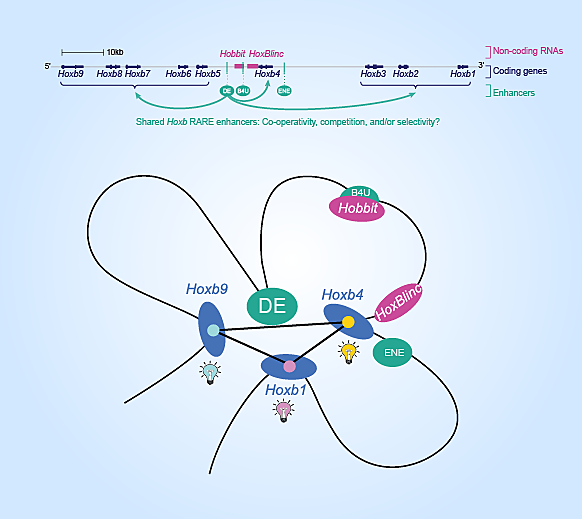
Hoxb1-9 genes are linearly located on a chromosome with three shared retinoic acid responsive enhancers situated in the middle, regulating their transcription (top). However, chromosome shape can bend, bringing enhancers to their target genes (bottom).
In a recent study published in Development on May 24, 2023, Zainab Afzal, Ph.D., a former University of Kansas Medical Center graduate student in the Krumlauf Lab, along with collaborators from the Stowers Technology Centers, uncovered the dynamic properties that three shared retinoic acid responsive enhancers have on regulating the transcription—the creation of gene messages to be translated into proteins—of the Hoxb gene cluster, which is responsible for specifying tissue identity along the head-to-tail axis of the mouse body plan.
The team sought to answer two questions. First, within single cells, how do shared, overlapping enhancers situated in the middle of a long stretch of DNA containing nine Hoxb genes coordinately regulate these genes over short and long distances? Second, do they turn on Hoxb genes simultaneously or one by one in one cell?
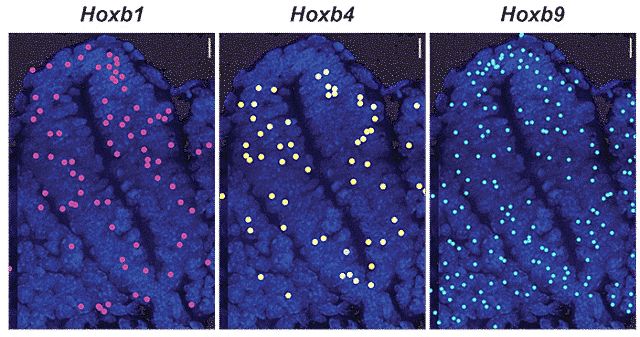
Transcription in real time within a mouse tissue neural tube cross-section for three Hoxb genes spanning the cluster.
The answers were surprising.
“It’s amazing what we don’t know.” – Robb Krumlauf
To answer if the enhancers impact Hoxb genes selectively or in coordination with each other, patterns of transcription for several of these genes spanning the entire cluster were analyzed. This was accomplished by taking snapshots of transcription activity at a single point in developmental time on several sections of mouse embryos.
Together, these three enhancers are required for controlling the correct level and activity of gene expression at precise points in time and space in the developing embryo. However, each of these regulatory elements have distinct tendencies, with two of the three having a more global role in regulating the Hoxb cluster while one has a more proximal preference for nearby genes.
By analyzing whole tissue sections at the single cell level, nearly all measurements indicated that only one gene was active at a time in a cell. Thus, Hoxb genes are not turned on simultaneously in a single cell, but rapidly and dynamically activated independently.
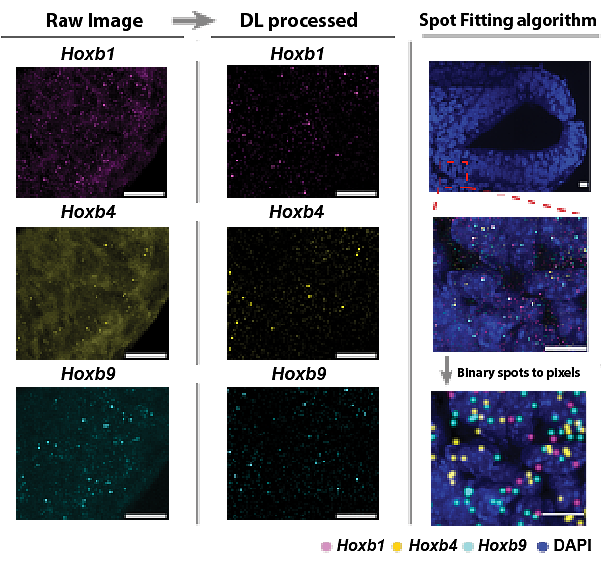
Technologies and techniques: Intron probes were developed for several coding and non-coding DNA regions. The raw images from nascent transcription of Hoxb1, 4, and 9 undergo deep learning processing and spot-fitting to count active transcription in single cells.
“These results suggest that the genes might be in a transcriptional hub with a 3D structure that positions them in close proximity to the enhancers despite their linear position on the chromosome,” said Krumlauf. “In hubs, the enhancers may be positioned to dart in and out to engage different target genes and rapidly coordinate gene activity for genes that are far apart on DNA.”
Ground Control to Major Tom: It’s time for a technology boost
The technologies and techniques developed and used in this study were integral for addressing these fundamental questions about transcription. The Krumlauf Lab in conjunction with several scientists from the Technology Centers, developed several fluorescent probes. Each probe was designed to emit a specific-colored spot when bound to actively transcribing RNA for three Hoxb genes spanning the entire cluster (Hoxb1, 4, and 9).
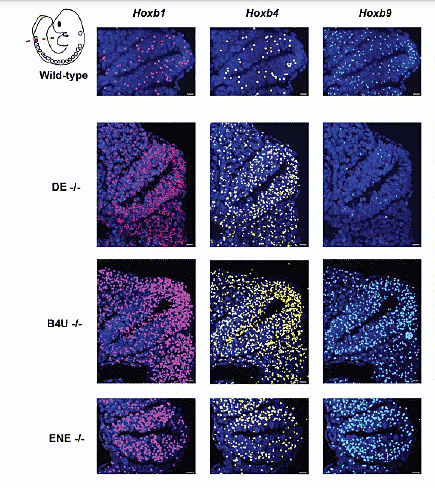
Each single enhancer mutation amplifies Hoxb gene expression compared to mice without enhancer mutations (wild-type).
Zooming in on the nucleus, the physical location of gene transcription in a cell, in different sections of a mouse embryo, the probe images were divided among several scientists for a manual fluorescent spot count. Using the results from each person’s tally to train a computer to “learn” this counting technique allowed the team to develop a deep learning algorithm that was just as accurate, if not better, than the manual method—and much faster.
Additionally, while the three enhancers were known to be required for Hoxb gene transcription, how each one contributes was addressed by making function-disrupting mutations for each, and then examining mouse lines harboring one, two, or all three mutations.
A result that the team did not expect was that single enhancer mutations actually increased gene activity across the Hoxb cluster. Even more astonishing was that transcription activity returned to expected levels when two of the three enhancers were mutated. Hoxb expression was null when all three were disrupted.
“The results from the single and double mutations were really surprising to us,” said Krumlauf. “Somehow the cooperativity between the enhancers was altered and it may be better to have neither than to have one without the other balancing it.”
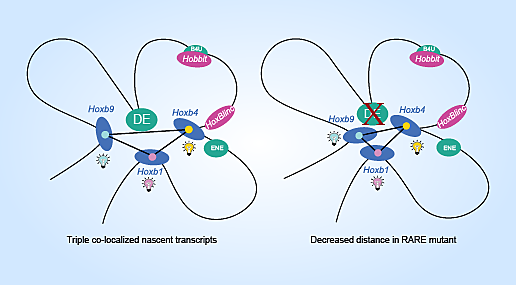
Single mutations may amplify transcription by acting to decrease the distance between target genes.
Space: the final frontier
Collective findings from the study indicate that the basic mechanisms of transcription that were thought to be understood are regulated in a much more complex way. Combining new technologies and rethinking previously presumed principles will provide a far greater understanding of how cell and tissue identity is determined in the right way, at the right time to build an organism.
NASA astrophysicist Jane Rigby told the audience at the 2023 American Association for the Advancement of Science annual meeting, “There are two ways to succeed in astronomy: either be really smart or be there at the right time.”
Krumlauf takes a strikingly similar stance on the study of transcription; a combination of intelligence and observation at the right time both factor in. “We are really trying to understand the dynamics of how transcription happens in real time, and the Technology Center experts were just as fascinated by these questions as we were,” said Krumlauf. “For me, whenever you have smart people interested in a problem, throwing out ideas and struggling through setbacks, you come up with a solution.”
Additional authors include Jeffery Lange, Ph.D., Christof Nolte, Ph.D., Sean McKinney, Ph.D., Christopher Wood, Ph.D., Ariel Paulson, Ph.D., Bony De Kumar, Ph.D., Jay Unruh, Ph.D., and Brian Slaughter, Ph.D.
This work was funded by institutional support from the Stowers Institute for Medical Research.
Read more here on Development's Research Highlight and an interview with Krumlauf and Afzal:
RARE peak into Hoxb transcriptional regulation
The people behind the papers - Robb Krumlauf and Zainab Afzal
In The News

07 January 2026
From the Journal of Cell Science, Investigator Kamena Kostova named a 'Cell Scientist to Watch'
Read Article
#Stowers25: Celebrating 25 Years
06 January 2026
Alejandro Sánchez Alvarado, Ph.D., reflects on a year of discovery, gratitude, and the community that helps support our mission.
Read Article
In The News
01 January 2026
From Science Friday, President and CSO Alejandro Sánchez Alvarado talks about the science of regeneration and the biology lessons we can carry into the new year.
Read Article
