By Marla Vacek Broadfoot
Navigating New Biological Terrain with Teamwork and Technology
During the Age of Exploration in the fifteenth and sixteenth centuries, a rush of European explorers set out to find fabled lands such as the gold and spice islands of Asia. Many of these sea travelers, however, embarked on their voyages under a number of misconceptions. The best map in the late fifteenth century indicated that Asia was much larger, and closer to Europe by water, than was actually true. When the explorers ran upon solid ground again—landing on an island perhaps, or the coast of North or South America—they often had no idea where they were. It wasn’t on the map.
Today, it is hard to imagine such a mind-bending revelation. Centuries of exploration and documentation have charted practically every mountain, valley, and waterway, and the recent introduction of global positioning system (GPS) satellites and smartphones has allowed us to zero in on any location, cruise down any street, peek into any yard. Yet there is plenty of uncharted territory left to explore. Out in the cosmos, astronomers are discovering new planets. Under the seas, marine biologists are identifying new species. Deep inside cells, biologists are mapping new structures and molecular interactions, with unprecedented precision.
Much like GPS revolutionized our understanding of geography, the advent of robotics and miniaturization is enabling scientists to see a part of ourselves that we might not have known existed. At the Stowers Institute, laboratory researchers are working side by side with scientific support teams to plot the unknown. It is an exciting time marked by technological sophistication and eye-opening insights. And it is moving at a dizzying pace.
“We can’t get enough,” says Alejandro Sánchez Alvarado, PhD, an investigator at the Institute and a frequent collaborator with the Institute’s scientific support groups. “It’s like discovering new terrain on a regular basis.”
A Fantastic Voyage
As the needs of the Institute’s research programs have emerged and evolved over the past decade and a half, Stowers scientific support groups have grown and adapted as well. Two of these groups—Histology and Electron Microscopy—have experienced several similar transformations in their parallel growth trajectories at the Institute. And, as is the norm for Stowers scientific support groups, their paths intersect regularly in the course of seeking answers to biological questions.
The fields of histology and electron microscopy have been around for a long time. Histology was developed in the mid-1800s, when scientists discovered that they could use a variety of dyes to stain different parts of cells—and cell types—in glittering shades of purple, pink, blue, gold, and silver. Electron microscopy came much later, in the 1930s. Instead of using light to image a specimen, electron microscopes use beams of electrons, which have much shorter wavelengths and thus can reveal the structure of smaller objects. Both methods were incredibly tedious and time-consuming, and over the decades, started to show their age. Then, at the turn of the last century, these relatively old technologies began to incorporate advances in computer science and automation, making them more valuable than ever before.
Tari Parmely came to the Stowers Institute in the midst of this renaissance. The native Kansan began working as a research specialist in the laboratory of Joan and Ron Conaway in 2002 and took over the management of the Tissue Culture Facility in 2008. In 2009, she added Media Prep (which prepares nutrient broths and agars for growing microorganisms) and in 2010 added Histology and Electron Microscopy.
Back then, the Histology Facility was staffed by several talented histotechnologists, who had been trained primarily in a hospital setting. But at the Institute, scientists don’t just deal with human tissue. They work with a multitude of model systems—flatworms, sea anemones, mice, yeast—and the list keeps growing. Every organism is different, and every tissue is unique. Each time a new model system comes online, the group has to develop or refine processing techniques to optimize the images that they produce for the researcher. So one of the first things Parmely set out to do as the head of Histology was to add research expertise to the lab.
Parmely found this expertise in Yongfu Wang, PhD, an assistant research professor at the University of Kansas who was well versed in both histology and research. “The rest of the team really welcomed him, and they have all worked so well together,” says Parmely. “It has been a beautiful thing to see. He has expanded the lab’s capabilities with molecular histology techniques, and that has helped the lab evolve into a more collaborative space.”
The Electron Microscopy Facility has also grown under Parmely’s stewardship. Initially, the facility consisted of a microscope, an ultramicrotome (the instrument used to cut specimens into thin slices for imaging), and Fengli Guo, PhD, a specialist to operate them both. The group has expanded to include three specialists and a technician, along with new equipment and technology. Today, the facilities can do much more than process samples—they can help researchers plot the course of their experiments, and provide the latest tools to ensure smooth sailing along the way.
Investigator Linheng Li, PhD, is interested in exploring the environment in and around tumors that allow cancer to relapse. For the last several years, he has focused his attention on a special type of cell known as the cancer stem cell, notorious for its ability to rebuild tumors decimated by chemotherapy and radiation. Xi (CiCi) He, MD, a senior research specialist in Li’s lab, worked with both the Histology and Electron Microscopy Facilities to examine tumors from a mouse model of colorectal cancer. She recalls the team being just as devoted to the project as she was. “They were very collaborative. Sometimes we finished an experiment late in the evening, after the staff had gone home, but they came back to fix the sample, so we could get it under the microscope quickly,” says He.
There, amid the dying cells, He witnessed an amazing rescue attempt undertaken by an unlikely ally: macrophages, the white blood cells that swarm to sites of infection and injury and gobble up cellular debris. The cells that were being killed off by chemotherapy and radiation sent out distress signals to macrophages, which in turn recruited cancer stem cells to produce more tumor cells. He showed that these signals could be blocked by a clinically approved drug called Celecoxib, that when combined with traditional cancer treatments could reduce tumor size and tumor number—and prevent the drugresistant cancer stem cells from spawning a relapse.
“It was an amazing discovery, and they made it together,” says Li. “The core facility members have ownership of the work, too.”
Intellectual explorers
With scientific knowledge accumulating at an exponential rate, researchers and specialists at the Institute have to make a concerted effort to keep up. Parmely feels it is important that members of scientific support teams hone their intellect by attending lab meetings, reading the literature, and participating in conferences. Some might attend conferences that feature advances in a specific technique, such as fluorescence in situ hybridization, whereas others might frequent larger gatherings that cover an entire discipline, like the National Society for Histotechnology annual meeting. The staff will pop in regularly to various meetings and seminars taking place within the halls of the Institute to find out what kinds of scientific questions are under investigation.
Ideas also bloom from conversations between researchers in different scientific support groups. Last year, Zulin Yu, PhD, a specialist in the Microscopy Center, which focuses on light microscopy techniques, mentioned to Wang in Histology a paper detailing a new technique called expansion microscopy that allows tiny structures to be visualized with nanoscale precision. Imagining how Stowers research projects might benefit from this approach, Yu and Wang set out to bring this technology to the Institute. Parmely recalls Wang tracking down and querying one of the inventors of expansion microscopy at a national meeting. “Yongfu was so excited about the technique,” she says. “He wanted to know every detail.”
Their determination and persistence paid off. With enabling contributions from a number of other Stowers colleagues in an impressive example of teamwork, Yu and Wang worked with Cori Cahoon, a predoctoral researcher in Investigator Scott Hawley’s lab, to apply the technique to a protein structure too small to be visualized by even the most high-power microscopes. That structure, known as the synaptonemal complex, is essential for the correct sorting of chromosomes into eggs and sperm during reproductive cell division or meiosis. The team gathered samples of the synaptonemal complex from dissected fruit fly ovaries, embedded the samples in a special type of gel, added liquid, and watched the samples expand fourfold. A key technical achievement by Wang was refining the sectioning procedure to allow the samples to be cut into thin slices prior to the fourfold expansion, thus permitting the use of advanced imaging techniques. When they put the samples under the microscope, they found that the structure looked like two sets of railroad tracks, one stacked on top of the other.
“The structure was so much more complicated and beautiful than we ever imagined. It completely changed the way we think about this structure and what it does,” says Hawley. “What they accomplished was a technical tour de force.”
New worlds
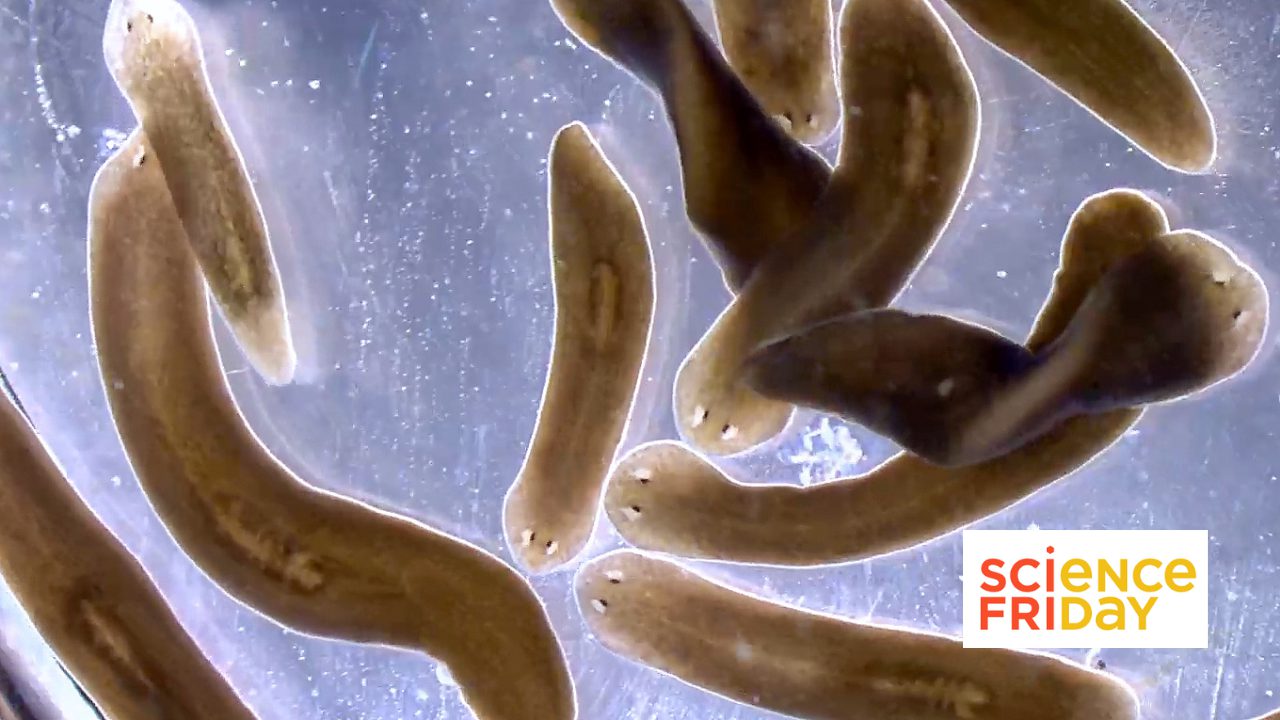
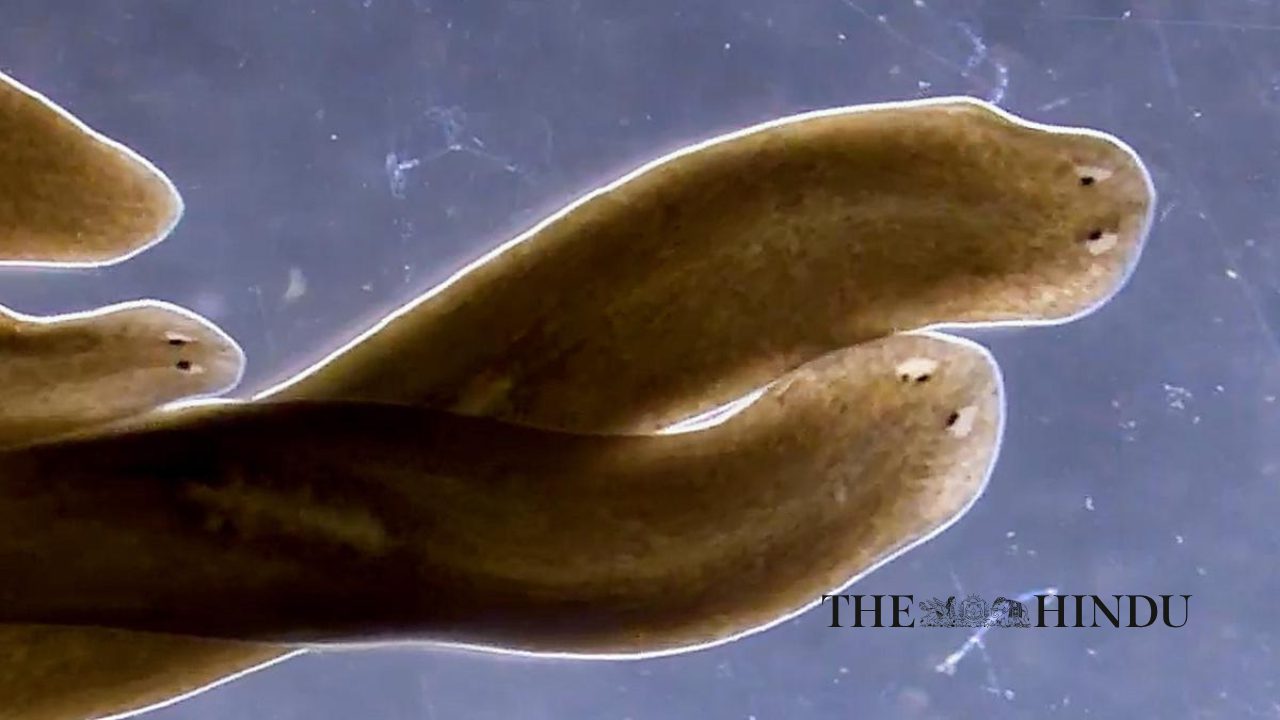
Mapping nano-sized structures and charting the interior of living organisms once seemed like impossible tasks. But recent technological advances are making the impossible possible. For twenty years, Sánchez Alvarado has studied planaria, arrow-shaped flatworms known for their amazing regenerative powers. These creatures embody an ideal system for understanding how some organisms regrow damaged organs or missing body parts. Sánchez Alvarado has long dreamed of digitally reconstructing the anatomy of a whole planarian, from tip to tip.
The feat—which entails dicing the miniscule worm into thousands of ultrathin sections, photographing each section individually, and then piecing the images back together again on the computer—would take a lifetime to complete with the old technology. Instead, his laboratory has resorted to analyzing a few small sections and then imagining how they might fit together to make the larger organism.
“It’s like sitting down to read James Joyce’s Finnegans Wake, and rather than tackling all two thousand pages, you just read every other hundred pages and assume from that sampling that you know more or less what the story is about. That’s no way to try to comprehend the whole body of work,” says Sánchez Alvarado.
Last year, Sánchez Alvarado learned about machines that could be adapted to section an entire planarian, process the sections, and stitch them together into a three-dimensional digital image. His dream project could become reality, attainable in a mere couple of years rather than a stretch of decades. Sánchez Alvarado wrote a grant to the Howard Hughes Medical Institute, which together with the Stowers Institute provided the funds to purchase the magical pieces of equipment, aptly named the MERLIN scanning electron microscope and the 3View ultramicrotome.
“For months and months, we were anticipating it arriving, and it was like Christmas morning when it showed up. It was like a big shiny toy,” says Parmely.
Right away, Stephanie Nowotarski, PhD, a postdoc in Sánchez Alvarado’s lab, began working with Melainia McClain and the Electron Microscopy team to get the machine online. Within a couple of months, it was up and running, and Nowotarski was snapping pictures of epithelium, the thin layer of tissue that covers the surface of the organism. The technology incorporates ATLAS.ti software, a sort of Google Earth app for electron microscopy that enables the user to zoom in and out on samples with varying degrees of magnification. When she shared her results at a lab meeting, she started by showing an image of a whole planarian worm, which is about the size of a toenail clipping. As she zoomed in further, she brought into focus the pores on its surface, and a gasp echoed throughout the room.
“It really is a team of equals here, and this collaboration opens the door to discovering new biology that we may not even know has been there under our noses the whole time,” says Sánchez Alvarado.
For example, Sánchez Alvarado says that scientists still don’t know how many cell types, or even how many individual cells, make up the human body. Is it 15 trillion? 70 trillion? What do they each look like, and how do they behave? By mapping the architecture of simpler systems like mice, fruit flies, and flatworms, researchers can set the course for creating a better understanding of the human condition.
After all, there is still much to be discovered, across the whole of biology. And science explorers could benefit from a better map.