In The News

07 January 2026
Investigator Kamena Kostova, named ‘Cell Scientist to Watch’
From the Journal of Cell Science, Investigator Kamena Kostova named a 'Cell Scientist to Watch'
Read Article
News
In zebrafish, three sequential phases are discovered to control hair cell regeneration

Zebrafish possess regenerative capacities that mammals do not. Sensory organs (neuromasts) are dotted along the zebrafish lateral line.
By Rachel Scanza, PhD
When was the last time you attended a rock show or an action movie in the theater? Well, let’s forgo the last two years, but in pre-pandemic life. How about the hands-clapped-over-your-ears discomfort of a fire or smoke alarm? Or a passing ambulance? Besides being loud, these commonplace events have sound intensities that can damage or destroy delicate cells in the ear that allow us to hear. We are born with a finite number of hair cells; repeated exposure to loud noise, certain medications, and aging results in permanent damage of these cells. The consequence? Hearing loss or deafness.
Species, like chicken, fish, and frogs, can regenerate, or create new cells or organs following an injury, an ability that mammals and in particular humans lack. Now, new research from the Stowers Institute for Medical Research details the molecular steps that enable zebrafish to create new sensory hair cells (HCs) when existing ones are destroyed. The study led by Postdoctoral Research Associate Sungmin Baek, PhD, from the lab of Tatjana Piotrowski, PhD, examines, at unprecedented resolution, the cellular mechanisms involved in HC regeneration. The findings published online in Developmental Cell on March 21, 2022, will aid in a more comprehensive understanding of regeneration with the potential to alter these, or very similar processes in different species including mammals.
“The idea is to identify molecular mechanisms that allow sensory organ regeneration in a regenerating animal,” said Piotrowski. “Hopefully from that we can learn where this process is blocked or not happening in mammals.”
Zebrafish possess sensory organs called neuromasts dotted along the lateral line of their body. Neuromasts are made up of different types of cells characterized by distinct genetic profiles. Within the neuromast are cells that detect water motion—termed “sensory hair cells” as they very closely resemble HCs in the human ear—and are surrounded by several types of support cells (SCs) that help maintain a sort of cellular equilibrium called homeostasis. When HCs are damaged or die, how are they replaced? SCs must change to replenish HCs, an orchestration both rapid and precise.

Stylized graphic illustrating the similarity between human hair cells and zebrafish sensory hair cells and the difference between regeneration and permanent loss following injury. Note: Over time, support cells actually populate gaps previously occupied by hair cells.
“The system is really ideal to study regeneration because sensory hair cells in the zebrafish lateral line regenerate extremely fast. This is one of the fastest regenerating cell types in a vertebrate described so far,” said Piotrowski.
Although an organ is composed of different cell types, the DNA in each of these cells is identical. What differentiates cell type, or cell gene expression, are the specific DNA sequences, or genes, that are transcribed into RNA messages. In this study, researchers use a technique called single cell RNA sequencing (scRNA-seq) to identify the activity of genes and cell types in HC regeneration at six, closely spaced time points.
Analyzing gene expression just minutes after inducing HC death and at five additional time points up to 10 hours reveals an exciting discovery; gene activity in SCs and in HC progenitors—a type of cell similar to a stem cell that maintains developmental gene programs—falls neatly into three unique time periods or modules where the preceding module activates the next one.
In the first module (0-30 minutes), immediately following HC death, many homeostatic genes are turned off while “injury response” genes are briefly activated. This time frame allows SCs to essentially “reprogram” their function and fate, and, would have gone unnoticed had researchers not collected data right away.
Between 30 minutes and one hour, or in module 2, while many gene activities are still inhibited, some genes referred to as “regeneration-specific” genes are temporarily activated. “What is interesting about these regeneration specific genes is that several of them are described in fin, retina and heart regeneration,” said Piotrowski. “It could well be that some of these genes or pathways are conserved between different organs.”
In the third module (3-10 hours) starting approximately three hours post HC death, researchers find a sudden upswing and reactivation of the programs that allow cells to regenerate. “We see a lot of genes suddenly turn on that are involved in transcription and protein translation that you would need to make hair cells during development,” said Piotrowski.
Another interesting discovery is that only a small subset of SCs actually become HCs with the rest returning to their original state, a decision occurring between three and five hours. “Somehow the cell senses that it has enough hair cells and suppresses further hair cell fate.”
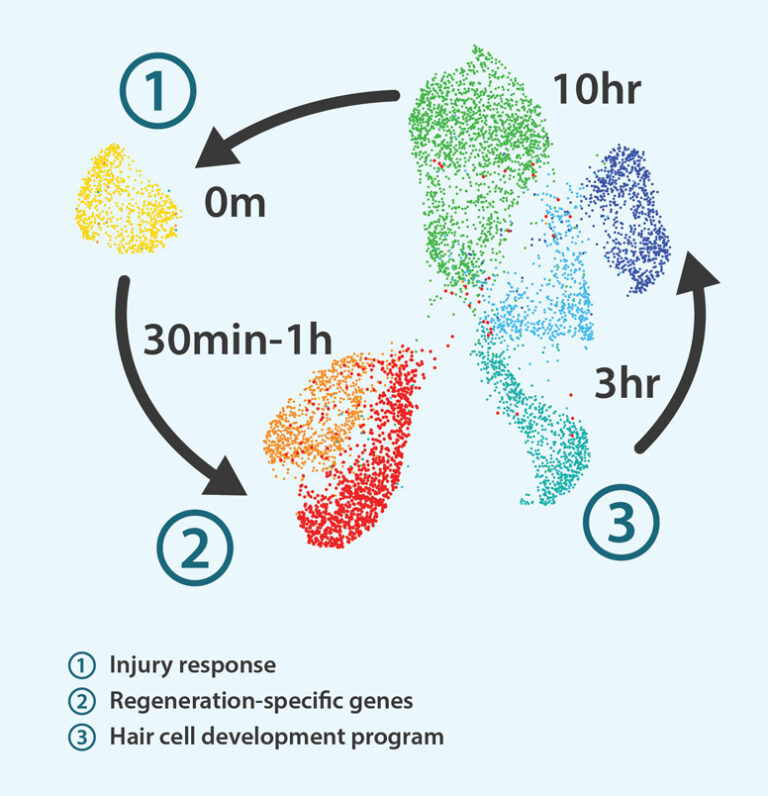
An atlas of regeneration showing gene expression changes over time. Each dot represents an individual central support cell (hair cell precursor), and colors represent cell collection times. Cells are grouped by similarity in transcriptional profiles, revealing three phases or modules of gene expression.
“The big question now is can we also find these three modules when we look at other regenerating organs,” said Piotrowski.
The researchers believe the initial inflammatory response activates regeneration but then needs to be suppressed for regeneration to proceed. The inhibition and subsequent enhancement of specific homeostatic genes is equally important; “downregulated” genes potentially could be temporarily turned off in mammals to aid in HC regeneration.
“The timing aspect of regeneration has not been described well so far,” said Piotrowski. “Knowing when to turn genes on and off and in which order can improve our understanding.”
The Hearing Health Foundation, of which Piotrowski is a member, aims to enhance our understanding of hearing loss via collaborative research to discover a cure. All data from the study is publicly available with the hope that scientists studying hearing loss and regeneration can use this to perform comparative studies across different organs and species including mammals.
“Our work provides a preliminary atlas of cell type and gene expression profiles. We hope to identify which genes are required to activate each module and to set a foundation for future studies focused on regeneration,” said Piotrowski.
Authors include Sungmin Baek, PhD, Nhung T. T. Tran, PhD, Daniel Diaz, Ya-Yin Tsai, Joaquín Navajas Acedo, PhD, Mark E. Lush, PhD, and Tatjana Piotrowski, PhD. This work was funded by the National Institute on Deafness and Other Communication Disorders of the National Institutes for Health (award 1R01DC015488-01A1) and by institutional support from the Stowers Institute for Medical Research. Data visualization and dissemination via gEAR was supported by grant R01DC019370.
In The News

07 January 2026
From the Journal of Cell Science, Investigator Kamena Kostova named a 'Cell Scientist to Watch'
Read Article
#Stowers25: Celebrating 25 Years
06 January 2026
Alejandro Sánchez Alvarado, Ph.D., reflects on a year of discovery, gratitude, and the community that helps support our mission.
Read Article
In The News
01 January 2026
From Science Friday, President and CSO Alejandro Sánchez Alvarado talks about the science of regeneration and the biology lessons we can carry into the new year.
Read Article
