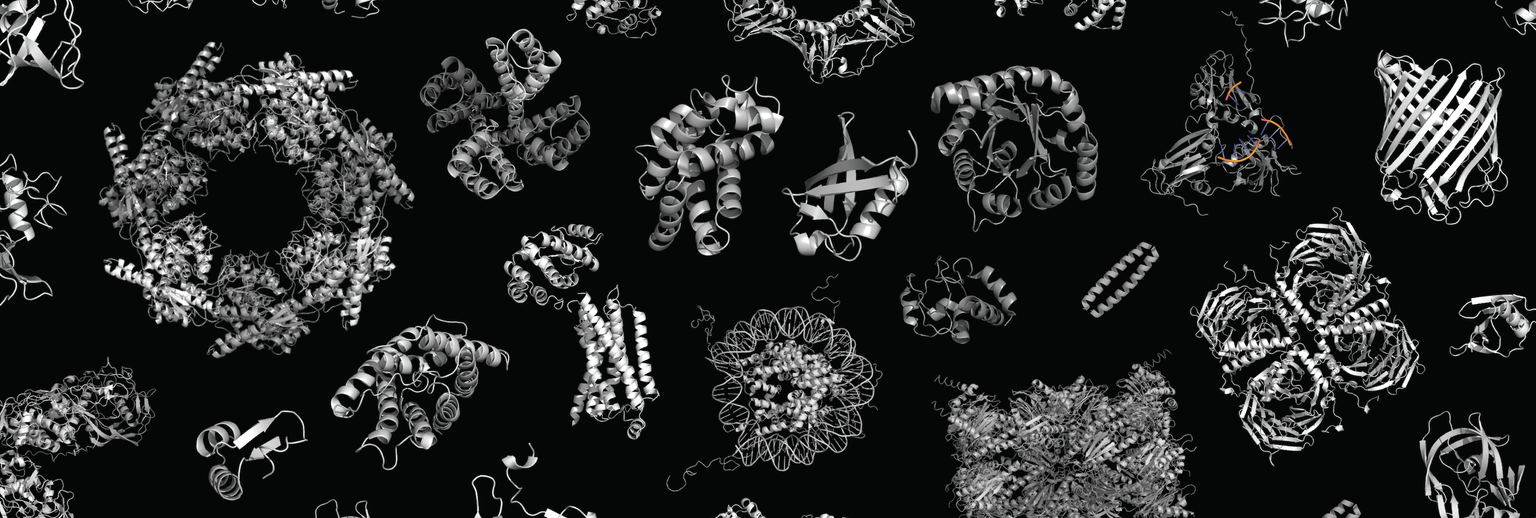
The Pillai Lab investigates the deep evolutionary origins of protein structure and function, and the biophysical principles that enable their design. The ability of proteins to fold, assemble into complexes, and carry out reactions required for life relies on precise atomic interactions between hundreds of individual atoms and amino acids. The molecular properties of proteins are the result of an evolutionary process stretching back 3.5 billion years. How does evolution – a blind stochastic process guided by natural selection – discover these sophisticated molecular solutions? What population genetic forces and biophysical constraints shape the evolution of complicated multi-residue protein features, like folding, assembly, catalysis, and allostery? What principles can we draw from evolution that could inform the design of synthetic proteins useful to biotechnology and medicine?
We combine evolutionary inference, computation, and experiments to test concrete, mechanistic hypotheses about the origins of molecular complexity. We resurrect ancestral proteins that bracket episodes of innovation in evolutionary history, use deep learning–guided structure prediction and design to evaluate evolutionary scenarios computationally, and express potential ancestral molecules in Escherichia coli. This allows us to quantify folding, interactions, catalysis, and conformational changes with a battery of biophysical, biochemical, and structural assays. Our questions – not a single model organism – define our scope, so we work across the tree of life, from viral accessory proteins and hemoglobin chaperones to spider-silk structural proteins.
We also aim to learn rules from evolution that could inform the design of synthetic proteins with biomedical purposes. Specifically, we are interested in building molecular machines - disaggregases and biosensors - that change structure in response to signals. In some cases, these machines convert chemical energy into mechanical work, opening the door to diagnostic biosensors and nanodevices that destroy disease-causing aggregates. More broadly, we hope to practically deploy protein design at the Stowers Institute to help address a range of biological questions in collaboration with other labs and Technology Centers.