In The News

07 January 2026
Investigator Kamena Kostova, named ‘Cell Scientist to Watch’
From the Journal of Cell Science, Investigator Kamena Kostova named a 'Cell Scientist to Watch'
Read Article
News
Stowers scientists identify “off” switches carried on some clusters of ribosomal RNA genes that can be passed from parent to child, a unique “fingerprint” of cluster size and activity that may help keep ribosomal RNA production in check.
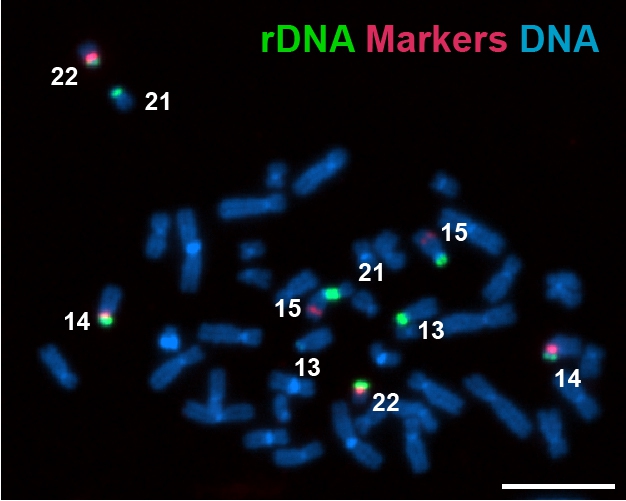
Fluorescent microscopy image of a chromosome spread identifying specific rDNA arrays.
For many years, scientists knew that the instructions for making ribosomal RNA (rRNA), a key component of ribosomes, are stored in large, repeated clusters or arrays on certain human chromosomes. But it wasn’t fully appreciated that these clusters come in different sizes on different chromosomes nor why some of these clusters are active while others are silent.
In a new study from the lab of Stowers Institute Investigator and Graduate School Dean Jennifer Gerton, Ph.D., in Cell Genomics on October 2, 2025, researchers revealed that some ribosomal DNA (rDNA) arrays are silenced or turned “off” by chemical tags on DNA while others remain active, a process called epigenetic regulation. These tags — tiny molecules called methyl groups — do not change the DNA sequence itself but attach to rDNA to influence whether rRNA production proceeds or not.
rRNA genes are typically present in excess copies — copies that can be passed from parents to their children — and this epigenetic silencing (via methylation of DNA) helps balance how many genes are active. The process ensures proper production of rRNA, a critical component of ribosomes.
“Everyone carries a distinctive rDNA fingerprint of array sizes and activity across chromosomes, and these on/off states can persist through development to guide proper ribosome production,” said Gerton.
"What surprised me most was that the epigenetic status of rDNA arrays can be inherited,” said Tamara Potapova, Ph.D., the study’s lead author and Research Specialist II in the Gerton Lab.“ In two family trios where the offspring had a silent array, we could trace it to the father, suggesting it propagated across generations.”
Ribosome production is the cell’s most energy-intensive job, and genomes carry far more rRNA genes than needed at any one time. Also, ribosomes are vital for all cell functions, and the correct number of active rRNA genes is important for cell function. Understanding these switches could potentially inform approaches to diseases involving disrupted ribosome formation and cell health.
Potapova explained, “This silencing process may act as a balancing system, keeping cells from making too many copies of rRNAs used to build ribosomes. It could also give scientists a way to explore how the environment shapes the activity of these genes. Additionally, the idea of each person having a unique rDNA “fingerprint” might one day be useful in areas like forensics science or tracing ancestry.”
By understanding what controls the epigenetic status of rRNA gene arrays, scientists can potentially test how environment might flip these switches and whether tuning them could affect a cell’s nucleolus structure and its ribosome production.
What they found
rRNA is the most abundant RNA in cells. Humans carry hundreds of rRNA genes arranged on chromosomes 13, 14, 15, 21, and 22. Using high-resolution imaging, long-read sequencing, and DNA-tag profiling, the team measured both the size of each chromosome’s rDNA array and whether it was actively making rRNA for 15 human genomes. They saw significant natural variation, showing that arrays can be large or small or sometimes even missing. Similar patterns were observed in great apes, suggesting this is a common feature found across humans and primates.
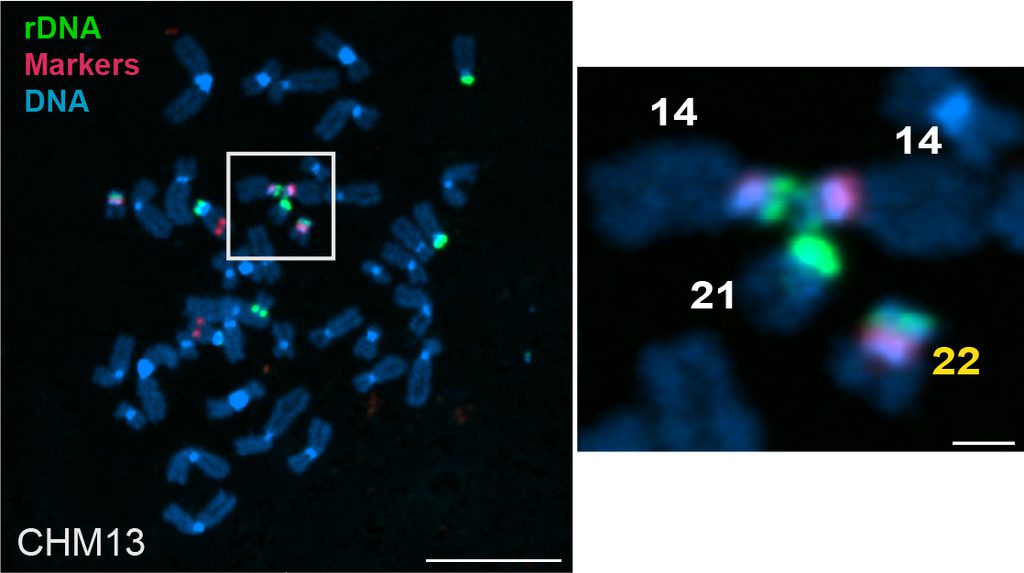
Fluorescent microscopy image of the chromosome spread from the CHM13 cell like showing rDNA linkages between acrocentric chromosomes (left). Magnification show linkage between two copies of chromosome 14 with a copy of chromosome 21 (right).
“This variation gives each of us a unique fingerprint of rDNA copy number and distribution,” said Potapova. “Because we inherit chromosomes from both parents — each with their own fingerprint — every person ends up with a different rDNA array size that may need to be finetuned.”
A deep learning pipeline
The study also showcases a technical leap: an image-analysis pipeline built from a trained deep learning model. “Tamara and Sean McKinney, Ph.D., Head of Computation Imaging trained a deep-learning model to analyze chromosome images at scale,” said Gerton. “Instead of scoring a handful of chromosomes by eye, we robustly quantified signals across thousands of arrays.”
This deep learning model characterizing chromosomes may be first of its kind. Gerton added, “Combined with computational calculations of rRNA gene copy number, the technique allowed us to assign each chromosome both a copy number and an activity level.”
Beyond genetics
When the researchers removed the chemical tags that keep genes off, the previously silent arrays became active and began producing their own distinct rRNA sequences — direct evidence that chemical tagging helps maintain the “off” state. Interestingly, the transmission of silent rDNA arrays were remarkably stable. Even when cells were reprogrammed into stem cells and grown into tiny brain and gut-like tissues called organoids, these arrays remained silent.
The findings raise questions about how much of the rDNA program survives the usual epigenetic “reset” between generations. Discovering these inherited fingerprints may have potential human health applications, for example, when rRNA production is dysregulated in human disease like cancer or ribosomopathies. It also advances our understanding of inheritance beyond just genetics.
Additional authors include Paxton Kostos, Matthew Borchers, Jeff Haug, Andrea Guarracino, Ph.D., Steven Solar, Mark Mattingly, Graciela Monfort Anez, Leonardo Gomes de Lima, Ph.D., Yan Wang, Ph.D., Chongbei Zhao, Ph.D., Kate Hall, Madelaine Gogol, Sophie Hoffman, Dmitry Antipov, Ph.D., Arang Rhie, Ph.D., Monika Sechova, Ph.D., Karen Miga, Ph.D., Erik Garrison, Ph.D., and Adam Phillippy, Ph.D.
This work was supported by the Intramural Research Program of the National Human Genome Research Institute, the National Cancer Institute of the National Institutes of Health (NIH) (award: R01CA2663393), and with institutional support from the Stowers Institute for Medical Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
In The News

07 January 2026
From the Journal of Cell Science, Investigator Kamena Kostova named a 'Cell Scientist to Watch'
Read Article
#Stowers25: Celebrating 25 Years
06 January 2026
Alejandro Sánchez Alvarado, Ph.D., reflects on a year of discovery, gratitude, and the community that helps support our mission.
Read Article
In The News
01 January 2026
From Science Friday, President and CSO Alejandro Sánchez Alvarado talks about the science of regeneration and the biology lessons we can carry into the new year.
Read Article
