News

31 March 2022
Completing the sequence
Stowers Institute researchers help assemble the complete human genome
Read Article
News
Collaboration with Stowers scientists reveals mechanism underlying a common chromosomal abnormality

Fluorescent image of chromosomes (blue) and centromeres (cyan), illustrating a Robertsonian translocation between chromosomes 14 and 21 (red).
By Rachel Scanza, Ph.D.
The Human Genome Project—the complete sequencing of an individual genome—was a monumental milestone in modern biology. Yet, to uncover what underlies our individuality, along with our similarities, a comprehensive understanding of human genetics requires a comparative approach from diverse datasets.
Enter the Human Pangenome Reference Consortium. New research published in Nature on May 10, 2023, assembled 94 human genomes to investigate natural genetic variation between humans. In the process of this work, a surprising discovery suggests how a common chromosomal abnormality implicated in infertility and congenital conditions occurs.
“The authors assembled nearly 100 complete human genomes selected from diverse geographies to enable us to understand human-to-human variations,” said co-author and Investigator Jennifer Gerton, Ph.D. from the Stowers Institute for Medical Research. “These assemblies revealed patterns of genetic variation across chromosome regions which were previously inaccessible, letting us answer a longstanding question about the most common kind of chromosomal abnormality in humans.”
Adam Phillippy, Ph.D., a collaborator at the National Human Genome Research Institute, added, “We did not anticipate this but it’s a beautiful example of how curiosity and collaboration can lead to significant insights for human health.”
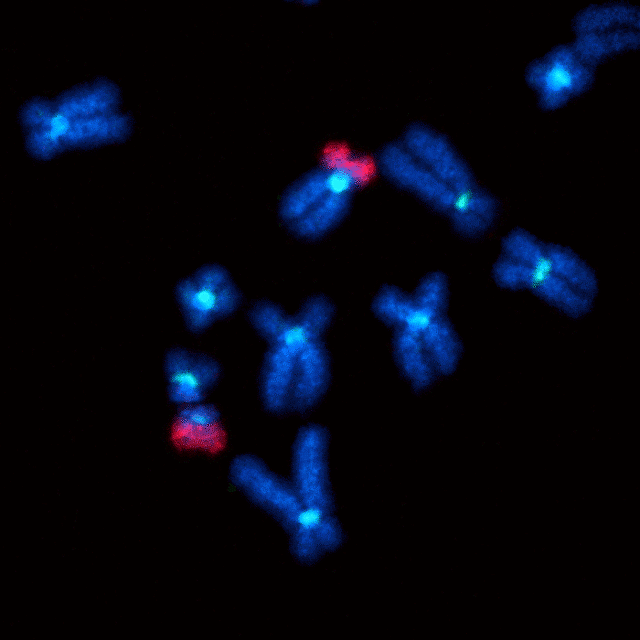
Closeup fluorescent image of a Robertsonian fusion between chromosomes 14 and 21 (red). There is one normal copy of chromosome 21 (upper center) and one with a fusion (lower left).
Insight into chromosome translocations
For the first time, analysis of assemblies from the Human Pangenome Reference Consortium reveals how and when specific translocations—a piece of one chromosome breaks off and attaches or fuses to another—called Robertsonian translocations, can form. Robertsonian translocations are the most common type of chromosomal fusion in the human population, occurring in one in 1,000 individuals, and contribute to infertility and genomic abnormalities like those that cause Down syndrome. The molecular basis for this type of translocation had until now remained elusive.
Humans have 23 pairs of chromosomes — the DNA/protein complex with two arms extending from the centromere, a region approximately at the center of the arms where duplicated chromosomes are held together prior to cell division. Five of the pairs do not look like the others. Termed human acrocentric chromosomes, these have asymmetric arm lengths, with one short arm and one long arm, compared with the remaining chromosomes. Of particular biological significance, the short arms of acrocentric chromosomes contain the genes required for the synthesis of ribosomes and ribosomal RNA. Ribosomes are required for manufacturing proteins, the workforce of a cell, and their formation is a cell’s most energy intensive activity.
During meiosis, a type of cell division that gives rise to sperm and eggs, genetic material can be exchanged or swapped between two paired chromosomes. This process, called recombination, where pairs of homologous chromosomes — one paternal and one maternal — break, and equal segments are swapped, increases genetic diversity in offspring because the chromosomes at the end of meiosis differ both from the parent and from each other.
An unexpected recombination result
However, analysis performed by the leader of the collaboration, Erik Garrison, Ph.D., of the University of Tennessee Health Science Center (UTHSC), revealed evidence for a different type of DNA exchange. Specific regions along the short arms of chromosomes 13, 14, 15, 21, and 22 are remarkably similar, indicating that recombination is occurring between chromosomes of mismatched pairs (for example, chromosomes 13 and 14 are exchanging information). The authors termed these “pseudohomologous” regions, to indicate that although they occur on different chromosomes, during meiotic recombination, they exchange sequences with other members of the acrocentric chromosome community as if they were homologs, or true pairs.

Human pangenome graph of the five acrocentric chromosomes with lines representing sequences with few variations and loops representing sequences with duplications and inversions. Acrocentric long arms (q) are separate while the short arms are shown in loops and tangles within dotted box.
These findings were achieved through the application of a novel bioinformatic approach. The team built a pangenome graph, which essentially acts as an alignment, or framework, to represent all possible similarities in sequence from dozens of genomes. Analyzing the graph’s structures enabled the researchers to infer the presence of regions on different chromosomes that are being homogenized, or made uniform, via recombination.
“We knew that there were regions of homology between the five acrocentrics, but we were amazed by the clarity and consistency of signals of recombination between heterologous acrocentric chromosomes in the pangenome,” said Garrison. “It was a eureka moment when we realized the same regions are involved in forming Robertsonian translocations,” added Gerton. “These regions are clearly undergoing frequent recombination—Robertsonian translocations represent a mistake in the process.”
Answering a 50-year-old mystery
Robertsonian translocations, first observed 50 years ago, result in the formation of a single chromosome when the long arms of two different acrocentric chromosomes fuse near their centromeres. The two short arms are lost amid repeated cell divisions and typically do not cause problems for their human hosts.

Graphical illustration of the mechanism by which recombination and Robertsonian translocation occurs between short arms of human acrocentric chromosomes. The physical proximity shown here between chromosomes 13 and 21 mediate recombination events (left and center). Robertsonian fusion (right) is illustrated between chromosomes 13 and 14.
However, individuals with this type of translocation are at greater risk for infertility and their offspring are more prone to trisomy conditions (having three copies of a chromosome instead of the usual pair) like Down syndrome, also known as Trisomy 21 (three copies of chromosome 21).
“Lots of people are Robertsonian carriers and don’t know it. Essentially you have a chromosome with an extra piece and during reproduction, this piece plus two normal chromosomes is similar to a trisomy,” said Gerton. “Inheriting the incorrect number of chromosomes, like Trisomy 21, increases with maternal age, however Robertsonian translocations are the most common cause of non-age associated Down syndrome.”
On chromosome 14, an inversion—a type of rearrangement where a chromosome segment breaks and rejoins in the opposite direction—in the pseudohomolous region relative to the other acrocentrics is strongly suspected as the culprit for Robertsonian translocation between chromosome 13 and 14, and 14 and 21, an idea the group plans to test in the future. Gerton explained, “This may ultimately lead to a better mechanistic understanding of what causes Down syndrome and other genetic disorders.”
The collaboration also paves the way toward considering these regions in biomedical and evolutionary studies that have previously overlooked them. “We haven’t been able to account for variation in the short arms of the acrocentrics in past studies of genome wide association and human evolution,” said Vincenza Colonna, Ph.D., a collaborator at UTHSC and the Institute of Genetics and Biophysics at the Italian National Research Council. “We show that these regions behave unusually genetically, and new approaches will be needed to leverage the information they contain into biomedical studies at the population level.”
Stowers co-authors include Boris Rubinstein, Ph.D., Leonardo Gomes de Lima, Ph.D., and Tamara Potapova, Ph.D.
Additional authors include Andrea Guarracino, Ph.D., Silvia Buonaiuto, Arang Rhie, Ph.D., Sergey Koren, Ph.D., Christian Fischer, and the Human Pangenome Reference Consortium.
This work was supported by the National Institute on Drug Abuse of the National Institutes of Health (NIH) (award: U01DA047638), the National Institute of General Medical Sciences of the NIH (award: R01GM123489), the Principles and Practice of Scalable Systems of the National Science Foundation (award: 2118709), the Tennessee Governor’s Chairs program, the Intramural Research Program of the National Human Genome Research Institute of the NIH, Human Technopole, Consiglio Nazionale delle Ricerche, Italy, and institutional support from the Stowers Institute for Medical Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
News

31 March 2022
Stowers Institute researchers help assemble the complete human genome
Read Article
News
31 March 2022
A ‘big data, AI, and genomics’ approach to identifying variation
Read Article
News
03 July 2019
Thanks to super-resolution microscopy, scientists have now been able to unambiguously identify physical associations between human chromosomes.
Read Article
