Press Release
21 February 2024
An awkward family reunion: Sea monsters are our cousins
Stowers scientists uncover how sea lamprey brain development is remarkably similar to that of humans
Read Article
We seek to understand the genetic and regulatory bases underlying body plan formation during development and how different species evolve.
Jump to:
Publications
Research Summary
Development and Regeneration, Genetics and Genomics, Evolutionary Biology, Molecular and Cell Biology, Neuroscience
Zebrafish, Mice, Fruit flies, Chicks, Sea lamprey
The Krumlauf Lab investigates the regulatory information and mechanisms that control the basic vertebrate body plan in development, disease, and evolution.
The lab focuses on head development and Hox genes—a family of genes that control the layout of a developing embryo from head to tail. The field offers insight into not only how bodies are shaped and formed, but also how species evolved new features. Fundamental discoveries in research organisms hold promise for understanding human health and disease.
The discovery that Hox genes, which control body plan formation, are essentially the same in mice and fruit flies (Drosophila) helped establish the idea that there are conserved genetic mechanisms. Comparative studies in mouse, chick, zebrafish, and sea lamprey continue to provide critical information on how different species form diverse structures.
Pioneering research from the Krumlauf Lab in the early 2000s also helped lay the groundwork for a new approach to treating osteoporosis — an often-debilitating disease that affects millions of people worldwide. The researchers uncovered a mechanism that controls bone growth, which arose out of their basic research into Hox genes. The drug, known as romosozumab, was approved by the U.S. Food and Drug Administration in January 2019, and is the first drug for osteoporosis that promotes bone growth.
The Krumlauf Lab continues to study the molecular and cellular pathways that govern the patterning of the nervous system, body plan, and craniofacial development of vertebrate embryos, particularly how these processes are altered or affected in human diseases.
Principal Investigator
Investigator and Scientific Director Emeritus
Stowers Institute for Medical Research
Robb Krumlauf, Ph.D., a world-renowned developmental biologist, joined the Stowers Institute in 2000 as the first Scientific Director and Investigator. He currently serves as an Investigator and Scientific Director Emeritus for the Institute and holds multiple faculty positions at the University of Kansas, the University of Kansas School of Medicine, and the University of Missouri-Kansas City Dental School.

Robb Krumlauf, Ph.D., a world-renowned developmental biologist, joined the Stowers Institute in 2000 as the first Scientific Director and Investigator. He currently serves as an Investigator and Scientific Director Emeritus for the Institute and holds multiple faculty positions at the University of Kansas, the University of Kansas School of Medicine, and the University of Missouri-Kansas City Dental School.
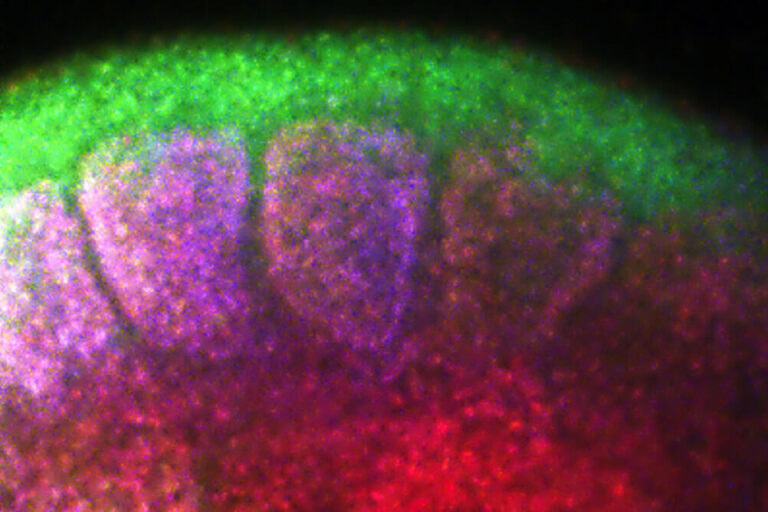
The Krumlauf Lab was one of the first to insert genes into the mouse genome to create transgenic mice that mimic human development. Krumlauf is also known for co-discovering collinearity of mammalian Hox genes, a phenomenon in which the order of these genes on the chromosome matches the order they’re expressed from head to tail along the embryo.
Press Release
21 February 2024
Stowers scientists uncover how sea lamprey brain development is remarkably similar to that of humans
Read Article
News
24 January 2024
The sea lamprey is a valuable research organism for exploring evolution
Read Article
News

01 July 2019
Over the ages, these classic song lyrics have taught countless children how their bodies are knitted together. But have you ever wondered why they fit together that way?
Read Article
News

08 July 2019
Pioneering research from the laboratory of Stowers Investigator Robb Krumlauf, PhD, helped lay the groundwork for a new approach to treating osteoporosis—an often debilitating disease that affects millions of people worldwide.
Read Article
Diversification and Functional Evolution of HOX Proteins
Singh NP, Krumlauf R. Front Cell Dev Biol. 2022;10:798812. doi: 798810.793389/fcell.792022.798812.
Transcriptional regulation and implications for controlling Hox gene expression
Afzal Z, Krumlauf R. J Dev Biol. 2022;10:4. doi: 10.3390/jdb10010004.
Singh NP, De Kumar B, Paulson A, Parrish ME, Scott C, Zhang Y, Florens L, Krumlauf R. [published ahead of print March 4 2021]. Dev Biol. 2021;9.
Segmentation and patterning of the vertebrate hindbrain
Krumlauf R, Wilkinson DG. Development. 2021;148,dev186460. doi:10.1242/dev.186460.
Parker HJ, De Kumar B, Pushel I, Bronner ME, Krumlauf R. Dev Biol. 2021:61-76.
A six-amino-acid motif is a major determinant in functional evolution of HOX1 proteins
Singh NP, De Kumar B, Paulson A, Parrish ME, Zhang Y, Florens L, Conaway JW, Si K, Krumlauf R. Genes Dev. 2020;34:1680-1696.
